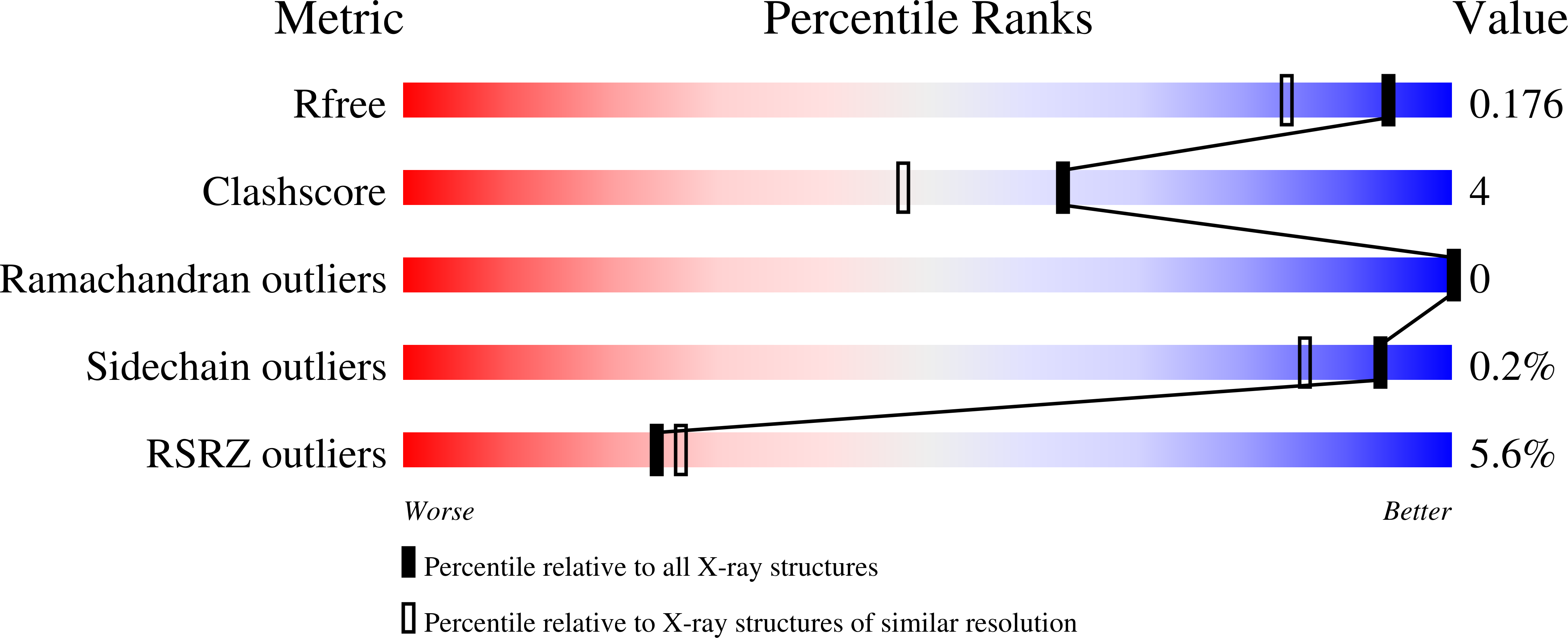
Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Lubner, C.E., Jennings, D.P., Mulder, D.W., Schut, G.J., Zadvornyy, O.A., Hoben, J.P., Tokmina-Lukaszewska, M., Berry, L., Nguyen, D.M., Lipscomb, G.L., Bothner, B., Jones, A.K., Miller, A.F., King, P.W., Adams, M.W.W., Peters, J.W.(2017) Nat Chem Biol 13: 655-659
- PubMed: 28394885
- DOI: https://doi.org/10.1038/nchembio.2348
- Primary Citation of Related Structures:
5JCA, 5JFC - PubMed Abstract:
The recently realized biochemical phenomenon of energy conservation through electron bifurcation provides biology with an elegant means to maximize utilization of metabolic energy. The mechanism of coordinated coupling of exergonic and endergonic oxidation-reduction reactions by a single enzyme complex has been elucidated through optical and paramagnetic spectroscopic studies revealing unprecedented features. Pairs of electrons are bifurcated over more than 1 volt of electrochemical potential by generating a low-potential, highly energetic, unstable flavin semiquinone and directing electron flow to an iron-sulfur cluster with a highly negative potential to overcome the barrier of the endergonic half reaction. The unprecedented range of thermodynamic driving force that is generated by flavin-based electron bifurcation accounts for unique chemical reactions that are catalyzed by these enzymes.
Organizational Affiliation:
Biosciences Center, National Renewable Energy Laboratory, Golden, Colorado, USA.