Crystal Structure of an Ammonia-Permeable Aquaporin.
Kirscht, A., Kaptan, S.S., Bienert, G.P., Chaumont, F., Nissen, P., de Groot, B.L., Kjellbom, P., Gourdon, P., Johanson, U.(2016) PLoS Biol 14: e1002411-e1002411
- PubMed: 27028365
- DOI: https://doi.org/10.1371/journal.pbio.1002411
- Primary Citation of Related Structures:
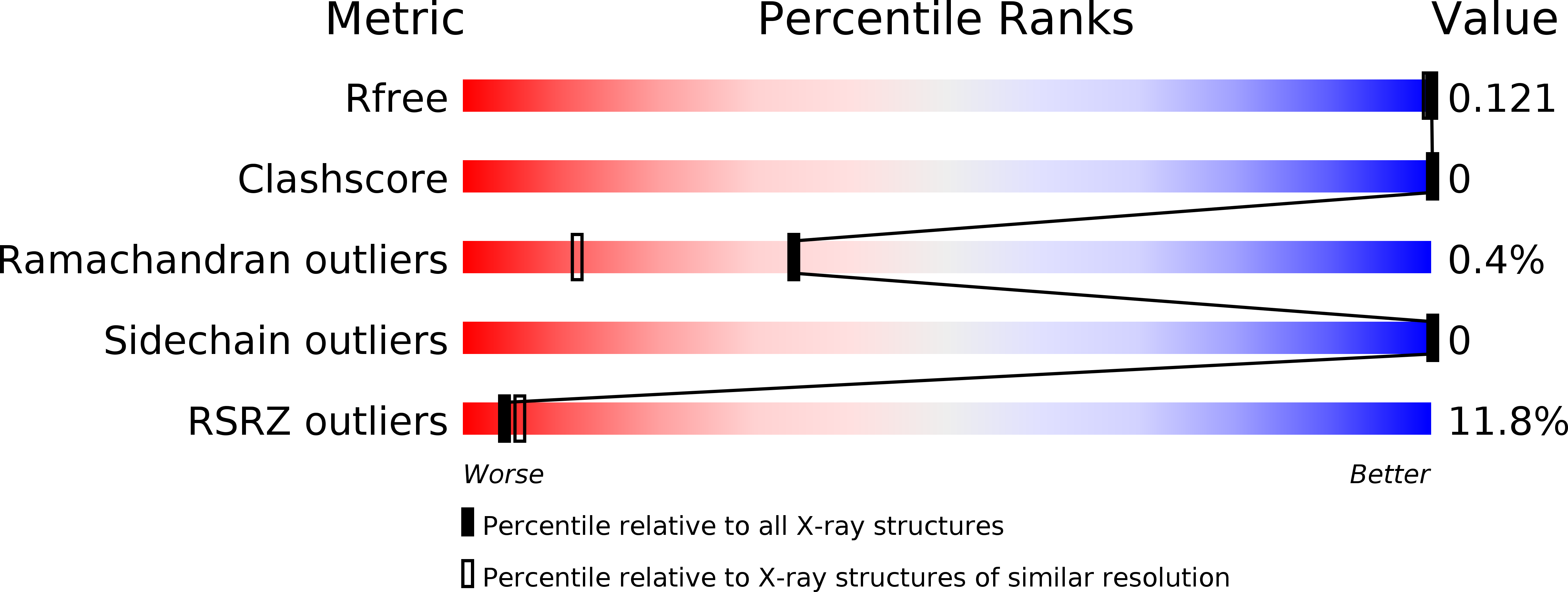
5I32 - PubMed Abstract:
Aquaporins of the TIP subfamily (Tonoplast Intrinsic Proteins) have been suggested to facilitate permeation of water and ammonia across the vacuolar membrane of plants, allowing the vacuole to efficiently sequester ammonium ions and counteract cytosolic fluctuations of ammonia. Here, we report the structure determined at 1.18 Å resolution from twinned crystals of Arabidopsis thaliana aquaporin AtTIP2;1 and confirm water and ammonia permeability of the purified protein reconstituted in proteoliposomes as further substantiated by molecular dynamics simulations. The structure of AtTIP2;1 reveals an extended selectivity filter with the conserved arginine of the filter adopting a unique unpredicted position. The relatively wide pore and the polar nature of the selectivity filter clarify the ammonia permeability. By mutational studies, we show that the identified determinants in the extended selectivity filter region are sufficient to convert a strictly water-specific human aquaporin into an AtTIP2;1-like ammonia channel. A flexible histidine and a novel water-filled side pore are speculated to deprotonate ammonium ions, thereby possibly increasing permeation of ammonia. The molecular understanding of how aquaporins facilitate ammonia flux across membranes could potentially be used to modulate ammonia losses over the plasma membrane to the atmosphere, e.g., during photorespiration, and thereby to modify the nitrogen use efficiency of plants.
Organizational Affiliation:
Department of Biochemistry and Structural Biology, Center for Molecular Protein Science, Lund University, Lund, Sweden.