KSHV SOX mediated host shutoff: the molecular mechanism underlying mRNA transcript processing.
Lee, H., Patschull, A.O.M., Bagneris, C., Ryan, H., Sanderson, C.M., Ebrahimi, B., Nobeli, I., Barrett, T.E.(2017) Nucleic Acids Res 45: 4756-4767
- PubMed: 28132029
- DOI: https://doi.org/10.1093/nar/gkw1340
- Primary Citation of Related Structures:
5HSW - PubMed Abstract:
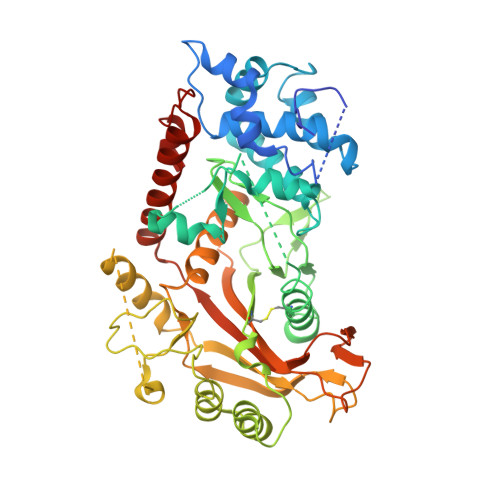
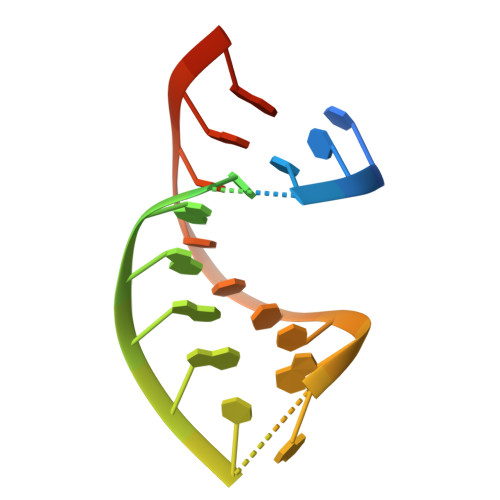
Onset of the lytic phase in the KSHV life cycle is accompanied by the rapid, global degradation of host (and viral) mRNA transcripts in a process termed host shutoff. Key to this destruction is the virally encoded alkaline exonuclease SOX. While SOX has been shown to possess an intrinsic RNase activity and a potential consensus sequence for endonucleolytic cleavage identified, the structures of the RNA substrates targeted remained unclear. Based on an analysis of three reported target transcripts, we were able to identify common structures and confirm that these are indeed degraded by SOX in vitro as well as predict the presence of such elements in the KSHV pre-microRNA transcript K12-2. From these studies, we were able to determine the crystal structure of SOX productively bound to a 31 nucleotide K12-2 fragment. This complex not only reveals the structural determinants required for RNA recognition and degradation but, together with biochemical and biophysical studies, reveals distinct roles for residues implicated in host shutoff. Our results further confirm that SOX and the host exoribonuclease Xrn1 act in concert to elicit the rapid degradation of mRNA substrates observed in vivo, and that the activities of the two ribonucleases are co-ordinated.
Organizational Affiliation:
Institute for Structural and Molecular Biology, Department of Biological Sciences, Birkbeck College, Malet Street, London WC1E 7HX, UK.