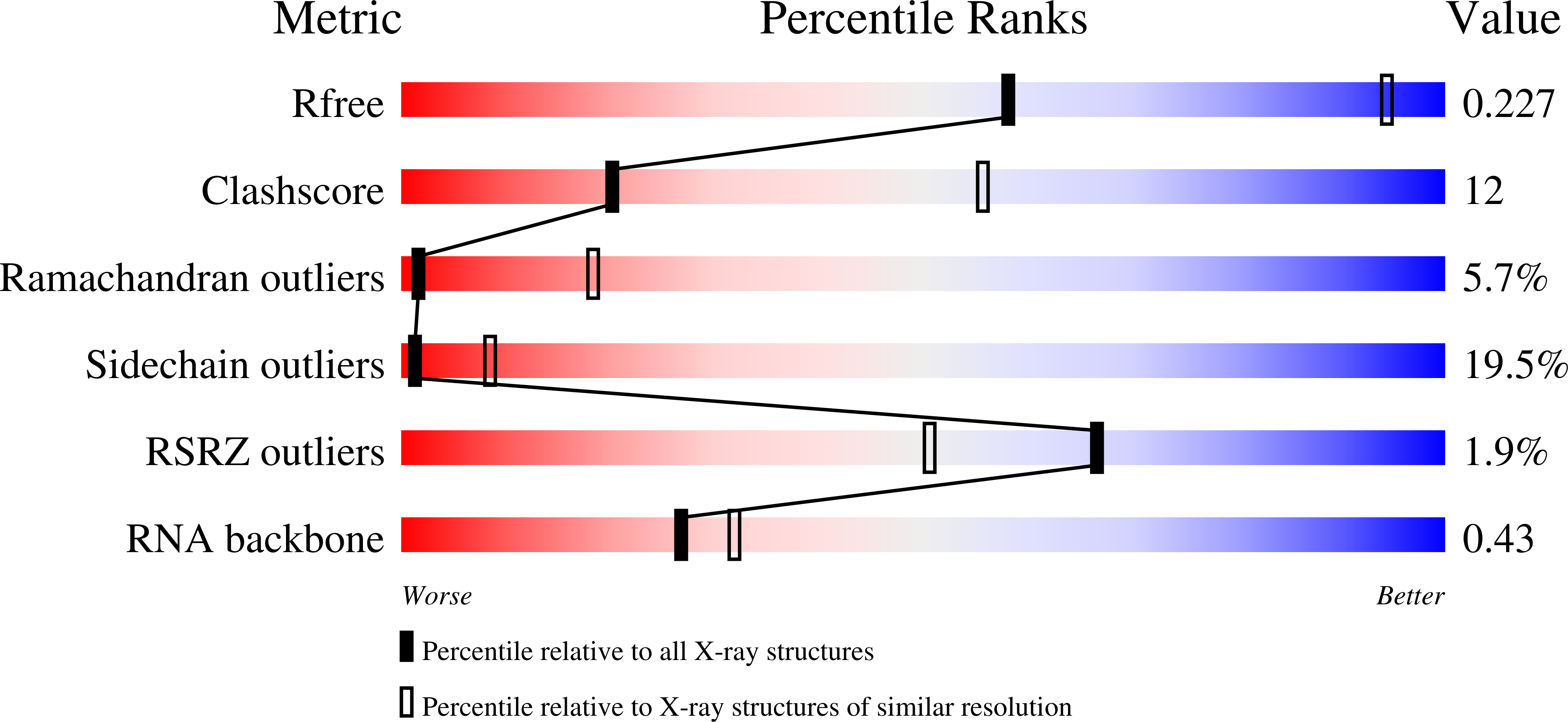
MN Query on MN Download Ideal Coordinates CCD File AB [auth X]
AB [auth X],
MANGANESE (II) ION MG Query on MG Download Ideal Coordinates CCD File AJ [auth X]
AJ [auth X],
MAGNESIUM ION