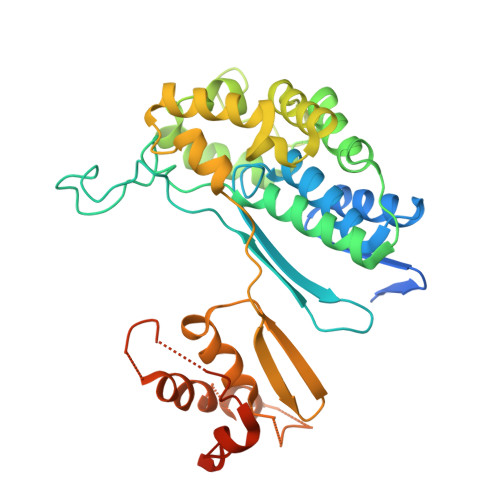
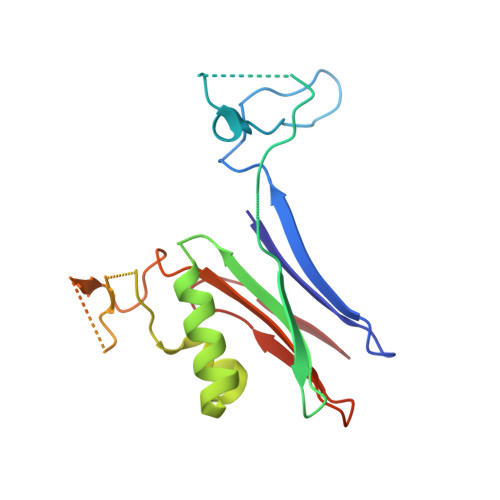
Biochemical and structural characterization of Klebsiella pneumoniae oxamate amidohydrolase in the uric acid degradation pathway.
Hicks, K.A., Ealick, S.E.(2016) Acta Crystallogr D Struct Biol 72: 808-816
- PubMed: 27303801
- DOI: https://doi.org/10.1107/S2059798316007099
- Primary Citation of Related Structures:
5HFT - PubMed Abstract:
HpxW from the ubiquitous pathogen Klebsiella pneumoniae is involved in a novel uric acid degradation pathway downstream from the formation of oxalurate. Specifically, HpxW is an oxamate amidohydrolase which catalyzes the conversion of oxamate to oxalate and is a member of the Ntn-hydrolase superfamily. HpxW is autoprocessed from an inactive precursor to form a heterodimer, resulting in a 35.5 kDa α subunit and a 20 kDa β subunit. Here, the structure of HpxW is presented and the substrate complex is modeled. In addition, the steady-state kinetics of this enzyme and two active-site variants were characterized. These structural and biochemical studies provide further insight into this class of enzymes and allow a mechanism for catalysis consistent with other members of the Ntn-hydrolase superfamily to be proposed.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA.