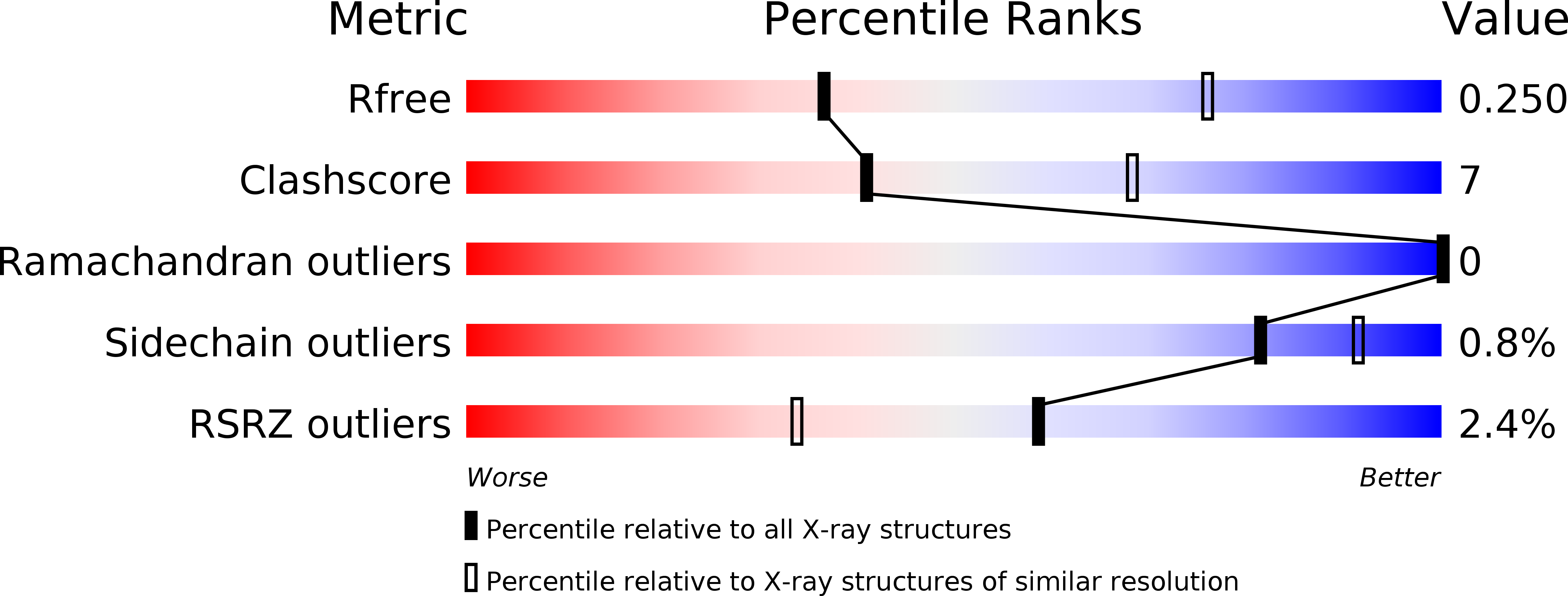
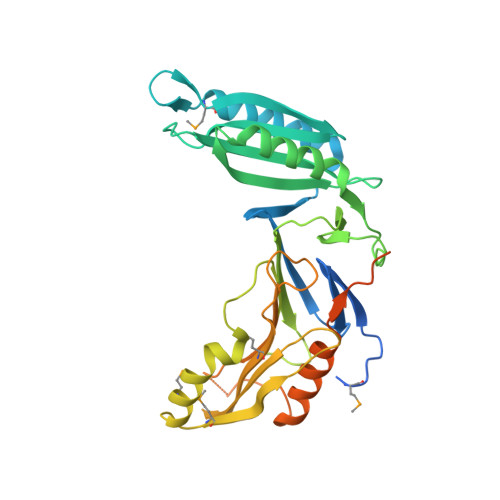
Structure-Based Prototype Peptides Targeting the Pseudomonas aeruginosa Type VI Secretion System Effector as a Novel Antibacterial Strategy
Gao, X.P., Mu, Z.X., Qin, B., Sun, Y., Cui, S.(2017) Front Cell Infect Microbiol 7: 411-411
- PubMed: 28979890
- DOI: https://doi.org/10.3389/fcimb.2017.00411
- Primary Citation of Related Structures:
5H7Y, 5H7Z - PubMed Abstract:
The type VI secretion system (T6SS) secretes numerous toxins for bacteria-bacteria competition. TplE is a newly identified trans-kingdom toxin secreted by the T6SS in Pseudomonas aeruginosa , while TplEi neutralizes the toxic effect of TplE to protect bacteria autointoxication. Blocking the interaction of TplE-TplEi could unleash the toxin, causing bacterial cell death. In this study, we applied a crystallographic approach to design a structural-based antimicrobial peptides targeting the interaction of TplE and TplEi. We found that a peptide (designed as "L" peptide based on its shape) derived from TplE can form a crystal complex with TplEi after subtilisin treatment and the crystal structure was solved at 2.2Å. The "L" peptide displays strong binding affinity to TplEi in vitro and can release the TplE toxin to induce bacteria death in vivo . Our findings suggest that as a toxin activator, the "L" peptide could be a possible drug lead for treating P. aeruginosa infection. Our findings provide an example that the T6SS effector and immunity protein could be a potential drug target against bacteria infection.
Organizational Affiliation:
MOH Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences and Peking Union Medical CollegeBeijing, China.