Structure and binding studies of proliferating cell nuclear antigen from Leishmania donovani.
Yadav, S.P., Singh, P.K., Sharma, P., Iqbal, N., Kaur, P., Sharma, S., Singh, T.P.(2017) Biochim Biophys Acta 1865: 1395-1405
- PubMed: 28844736
- DOI: https://doi.org/10.1016/j.bbapap.2017.08.011
- Primary Citation of Related Structures:
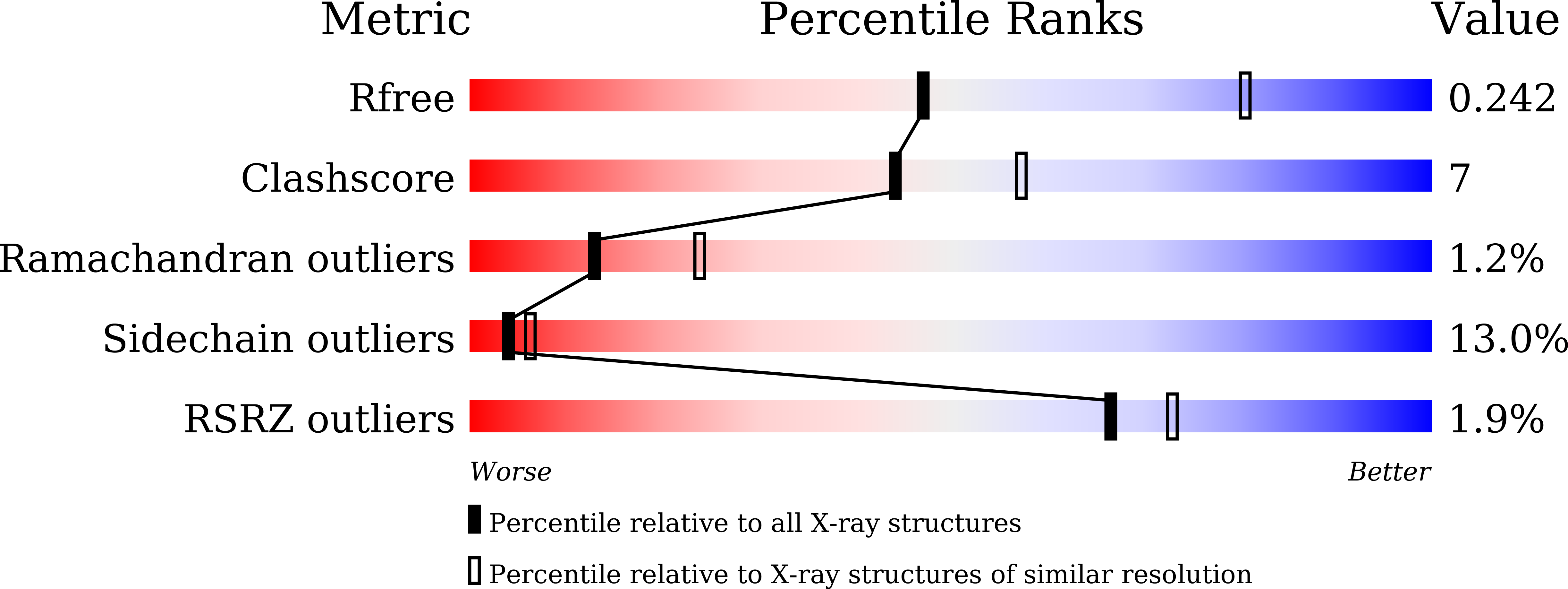
5H0T - PubMed Abstract:
Proliferating cell nuclear antigen (PCNA) acts as a sliding clamp to support DNA replication and repair. The structure of PCNA from Leishmania donovani (LdPCNA) has been determined at 2.73Å resolution. Structure consists of six crystallographically independent molecules which form two trimeric rings. The pore diameter of the individual trimeric ring is of the order of 37Å. The two rings are stacked through their front to front faces. In order to gain a stable packing, the rings are rotated by 42° about the pore axis and shifted by 7Å and tilted by 16° along the perpendicular direction to pore axis. This form of stacking reduced the effective diameter of the pore to 32Å. The sequence of LdPCNA consists of a long segment of 41 amino acid residues (186-Gly-Val-Ser-Asp-Arg-Ser-Thr-Lys-Ser-Glu-Val-Lys-Ala-Glu-Val-Lys-Ala-Glu-Ala-Arg-Asp-Asp-Asp-Glu-Glu-Pro-Leu-Ser-Arg-Lys-Tyr-Gly-Lys-Ala-Asp-Ser-Ser-Ala-Asn-Ala-Ile-226) whereas the corresponding segments in other PCNAs contain only eight residues corresponding to 186-Gly-Val-Ser-Asp-Arg------224-Asn-Ala-Ile-226. The enhanced length of this segment in LdPCNA may influence its mode of interaction with DNA and other proteins. The dissociation constants obtained using real time binding studies with surface plasmon resonance (SPR) for two peptides, Lys-Arg-Arg-Gln-Thr-Ser-Met-Thr-Asp-Phe-Tyr-His (P1) from human cyclin-dependent kinase inhibitor-1(CKI-1) and Lys-Thr-Gln-Gly-Arg-Leu-Asp-Ser-Phe-Phe-Thr-Val (P2) from flap endonuclease 1 (Fen-1) as well as with two small molecule inhibitors, (S)-4-(4-(2-amino-3-hydroxypropyl)-2, 6-diiodophenoxy) phenol hydrochloride (ADPH) and N-(3-methylthiophene-2-carboxylicacid)-N'-((3-hydroxy-2-naphthalenyl) methylene) hydrazide (MCMH) are 0.29±0.09μM, 0.37±0.08μM, 0.35±0.09μM and 1.20±0.08μM respectively. The corresponding values obtained using fluorescence spectroscopic methods were 0.22±0.06μM, 0.68±0.07μM, 0.44±0.07μM and 0.75±0.05μM respectively.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.