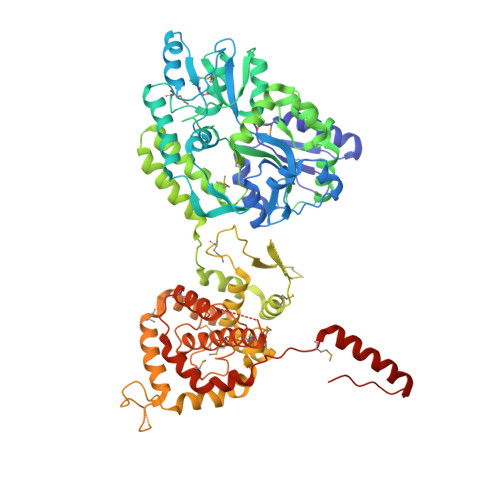
Structure of the Hantavirus Nucleoprotein Provides Insights Into the Mechanism of RNA Encapsidation.
Olal, D., Daumke, O.(2016) Cell Rep 14: 2092
- PubMed: 26923588
- DOI: https://doi.org/10.1016/j.celrep.2016.02.005
- Primary Citation of Related Structures:
5FSG - PubMed Abstract:
Hantaviruses are etiological agents of life-threatening hemorrhagic fever with renal syndrome and hantavirus cardiopulmonary syndrome. The nucleoprotein (N) of hantavirus is essential for viral transcription and replication, thus representing an attractive target for therapeutic intervention. We have determined the crystal structure of hantavirus N to 3.2 Å resolution. The structure reveals a two-lobed, mostly α-helical structure that is distantly related to that of orthobunyavirus Ns. A basic RNA binding pocket is located at the intersection between the two lobes. We provide evidence that oligomerization is mediated by amino- and C-terminal arms that bind to the adjacent monomers. Based on these findings, we suggest a model for the oligomeric ribonucleoprotein (RNP) complex. Our structure provides mechanistic insights into RNA encapsidation in the genus Hantavirus and constitutes a template for drug discovery efforts aimed at combating hantavirus infections.
Organizational Affiliation:
Crystallography, Max Delbrück Center for Molecular Medicine, Robert-Rössle-Strasse 10, 13125 Berlin, Germany. Electronic address: daniel.olal@mdc-berlin.de.