Biochemical and structural characterization of a keratin-degrading M32 carboxypeptidase from Fervidobacterium islandicum AW-1
Lee, Y.-J., Dhanasingh, I., Ahn, J.-S., Jin, H.-S., Choi, J.M., Lee, S.H., Lee, D.-W.(2015) Biochem Biophys Res Commun 468: 927-933
- PubMed: 26603937
- DOI: https://doi.org/10.1016/j.bbrc.2015.11.058
- Primary Citation of Related Structures:
5E3X - PubMed Abstract:
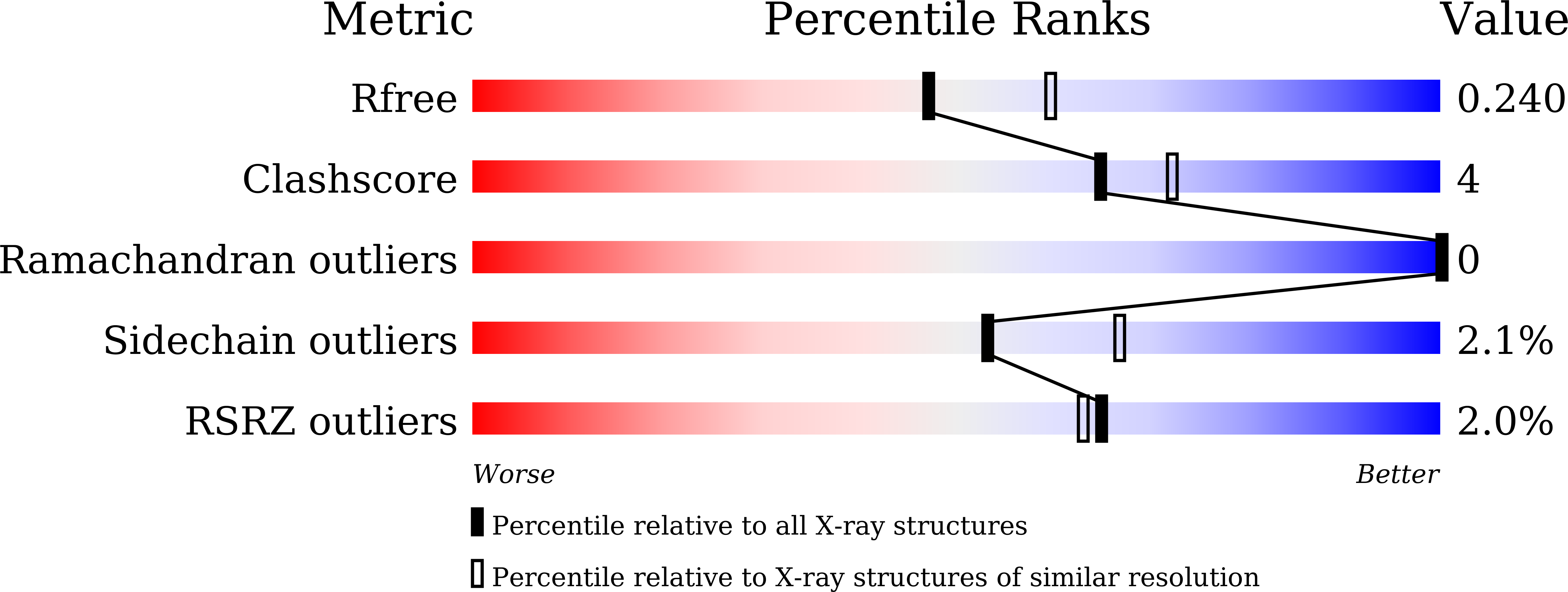
Comparative genomics of the keratin-degrading extremophilic eubacterium Fervidobacterium islandicum AW-1 and the closely related Fervidobacterium nodosum with no keratinolytic activity suggested that the FIAW1_1600 gene encoding a carboxypeptidase (CP) plays an important role in keratin degradation. The presumptive 489 amino acid sequence of the gene showed a conserved HEXXH motif with low levels of sequence identity (<38%) to reported thermostable M32 CPs. To identify its functional role, the FIAW1_1600 gene was overexpressed in Escherichia coli, and the recombinant enzyme was purified and characterized in detail. F. islandicum AW-1 CP (FisCP) formed a homodimer with a molecular mass of 107 kDa, and its apoenzyme exhibited maximal activity at 80 °C and pH 7.0 in the presence of Co(2+). This metalloenzyme mainly cleaved the C-termini of peptides with a basic amino acid sequence. The crystal structure of FisCP at 2.2 Å resolution showed high levels of structural similarities (root-mean-square deviations of <1.7 Å) to those of other M32 CP homologs. Remarkably, the enzyme significantly enhanced the degradation of native chicken feathers. This study suggests that FisCP, a keratinolytic member of the thermostable M32 CP family, plays an important role in keratin degradation for cellular metabolism in F. islandicum AW-1.
Organizational Affiliation:
School of Applied Biosciences, Kyungpook National University, Daegu 702-701, South Korea.