Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
Shimamoto, Y., Hattori, H., Kobayashi, K., Teruya, K., Sanjho, A., Nakagawa, A., Yamashita, E., Akaji, K.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 3C-like proteinase | 306 | Severe acute respiratory syndrome-related coronavirus | Mutation(s): 1 EC: 3.4.22.69 | 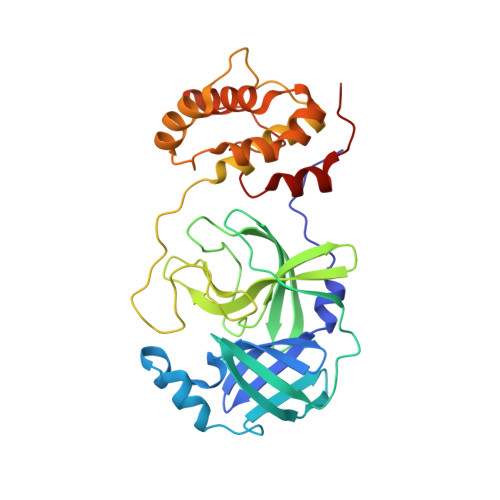 | |
UniProt | |||||
Find proteins for P0C6X7 (Severe acute respiratory syndrome coronavirus) Explore P0C6X7 Go to UniProtKB: P0C6X7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C6X7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SLH Query on SLH | B [auth A] | (2S)-3-(1H-imidazol-5-yl)-2-({[(3R,4aS,8aR)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal C25 H35 N5 O2 HCMRRQNNHUOUJM-NVXDSCFRSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 107.801 | α = 90 |
| b = 82.091 | β = 104.35 |
| c = 53.266 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DENZO | data collection |
| SCALEPACK | data reduction |
| PDB_EXTRACT | data extraction |
| MOLREP | model building |
| Coot | model building |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| MOLREP | phasing |