Design of dinuclear manganese cofactors for bacterial reaction centers.
Olson, T.L., Espiritu, E., Edwardraja, S., Simmons, C.R., Williams, J.C., Ghirlanda, G., Allen, J.P.(2016) Biochim Biophys Acta 1857: 539-547
- PubMed: 26392146
- DOI: https://doi.org/10.1016/j.bbabio.2015.09.003
- Primary Citation of Related Structures:
5C39 - PubMed Abstract:
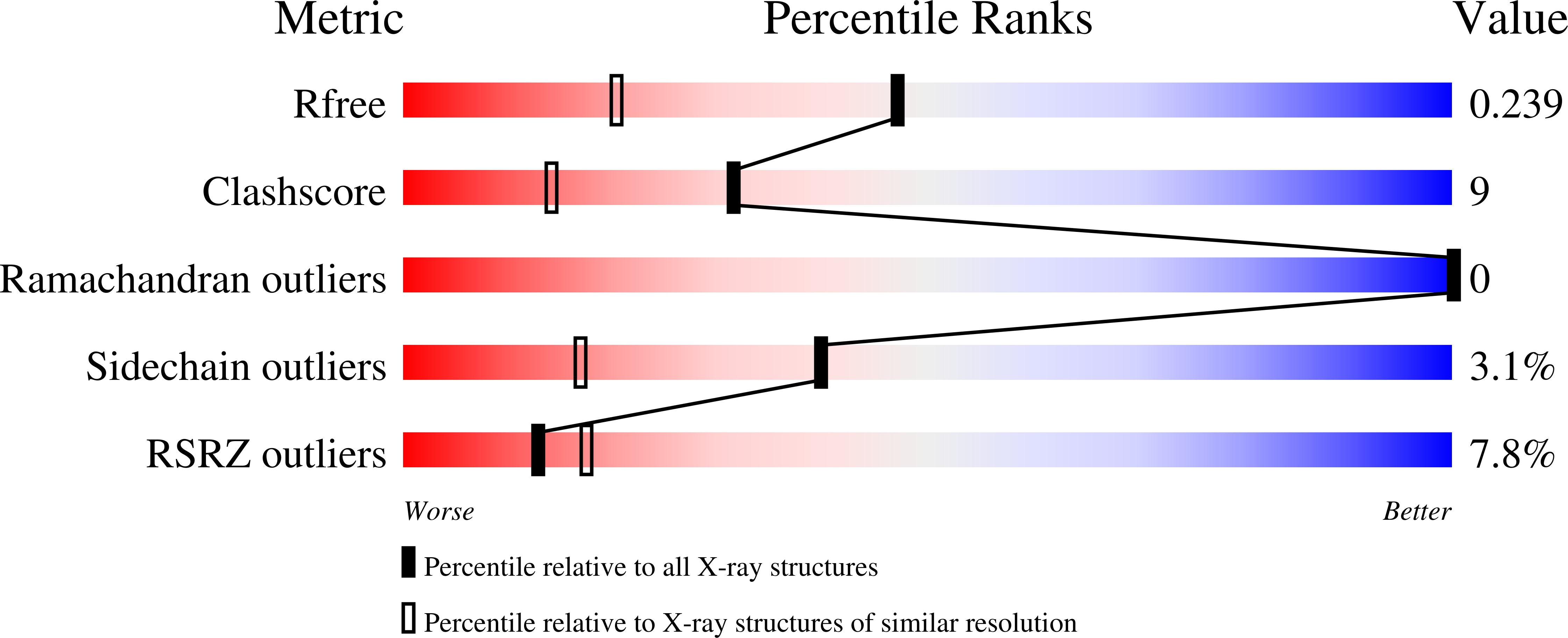
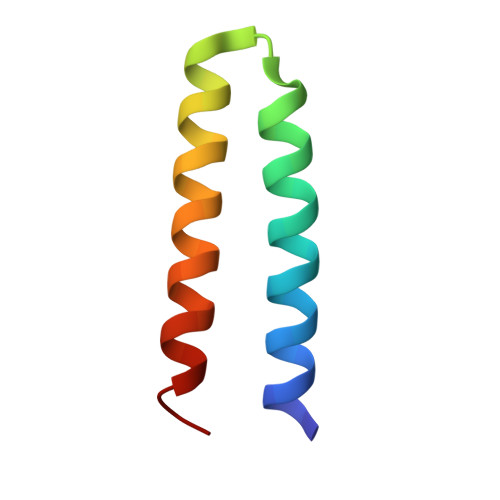
A compelling target for the design of electron transfer proteins with novel cofactors is to create a model for the oxygen-evolving complex, a Mn4Ca cluster, of photosystem II. A mononuclear Mn cofactor can be added to the bacterial reaction center, but the addition of multiple metal centers is constrained by the native protein architecture. Alternatively, metal centers can be incorporated into artificial proteins. Designs for the addition of dinuclear metal centers to four-helix bundles resulted in three artificial proteins with ligands for one, two, or three dinuclear metal centers able to bind Mn. The three-dimensional structure determined by X-ray crystallography of one of the Mn-proteins confirmed the design features and revealed details concerning coordination of the Mn center. Electron transfer between these artificial Mn-proteins and bacterial reaction centers was investigated using optical spectroscopy. After formation of a light-induced, charge-separated state, the experiments showed that the Mn-proteins can donate an electron to the oxidized bacteriochlorophyll dimer of modified reaction centers, with the Mn-proteins having additional metal centers being more effective at this electron transfer reaction. Modeling of the structure of the Mn-protein docked to the reaction center showed that the artificial protein likely binds on the periplasmic surface similarly to cytochrome c2, the natural secondary donor. Combining reaction centers with exogenous artificial proteins provides the opportunity to create ligands and investigate the influence of inhomogeneous protein environments on multinuclear redox-active metal centers. This article is part of a Special Issue entitled Biodesign for Bioenergetics--the design and engineering of electronic transfer cofactors, proteins and protein networks, edited by Ronald L. Koder and J.L. Ross Anderson.
Organizational Affiliation:
School of Molecular Sciences, Arizona State University, Tempe, AZ 85287-1604, USA.