Crystallographic and Computational Analyses of AUUCU Repeating RNA That Causes Spinocerebellar Ataxia Type 10 (SCA10).
Park, H., Gonzalez, A.L., Yildirim, I., Tran, T., Lohman, J.R., Fang, P., Guo, M., Disney, M.D.(2015) Biochemistry 54: 3851-3859
- PubMed: 26039897
- DOI: https://doi.org/10.1021/acs.biochem.5b00551
- Primary Citation of Related Structures:
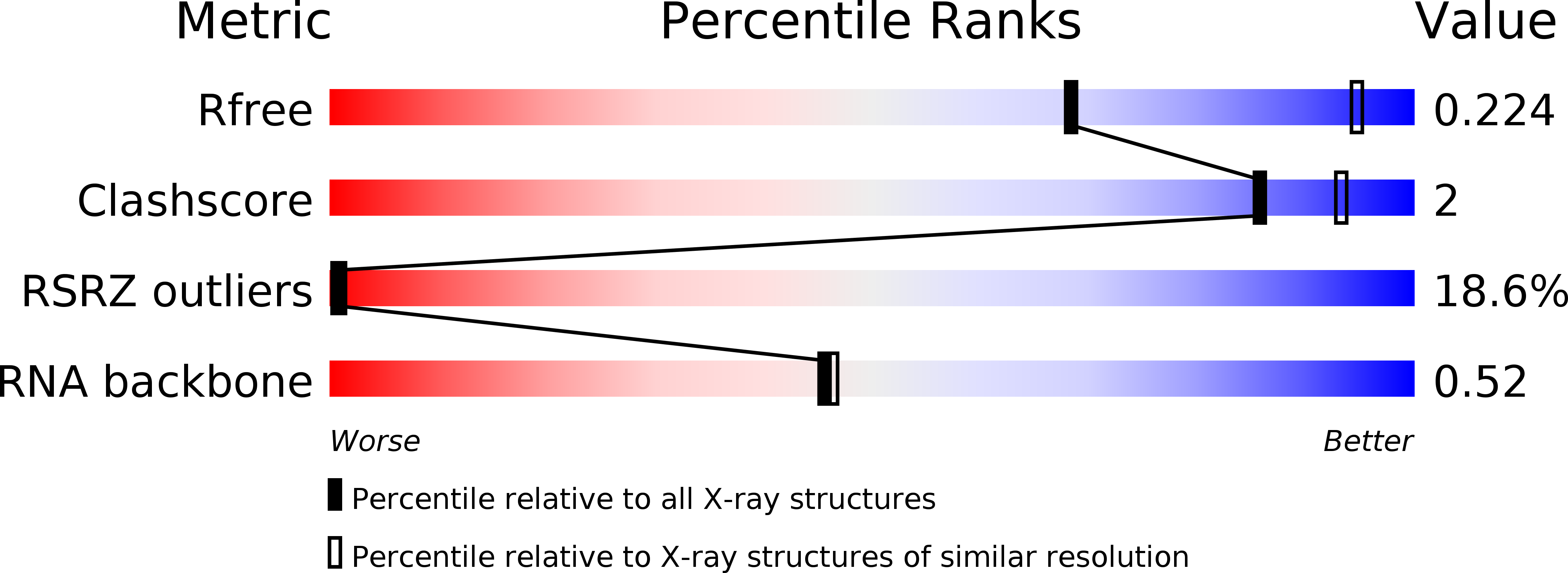
5BTM - PubMed Abstract:
Spinocerebellar ataxia type 10 (SCA10) is caused by a pentanucleotide repeat expansion of r(AUUCU) within intron 9 of the ATXN10 pre-mRNA. The RNA causes disease by a gain-of-function mechanism in which it inactivates proteins involved in RNA biogenesis. Spectroscopic studies showed that r(AUUCU) repeats form a hairpin structure; however, there were no high-resolution structural models prior to this work. Herein, we report the first crystal structure of model r(AUUCU) repeats refined to 2.8 Å and analysis of the structure via molecular dynamics simulations. The r(AUUCU) tracts adopt an overall A-form geometry in which 3 × 3 nucleotide (5')UCU(3')/(3')UCU(5') internal loops are closed by AU pairs. Helical parameters of the refined structure as well as the corresponding electron density map on the crystallographic model reflect dynamic features of the internal loop. The computational analyses captured dynamic motion of the loop closing pairs, which can form single-stranded conformations with relatively low energies. Overall, the results presented here suggest the possibility for r(AUUCU) repeats to form metastable A-from structures, which can rearrange into single-stranded conformations and attract proteins such as heterogeneous nuclear ribonucleoprotein K (hnRNP K). The information presented here may aid in the rational design of therapeutics targeting this RNA.
Organizational Affiliation:
∥Grup d'Enginyeria Molecular (GEM), Institut Químic de Sarrià (IQS)-Universitat Ramon Llull (URL), Barcelona 08017, Spain.