Inhibitor bound in the active site of Mycobacterium tuberculosis anthranilate phosphoribosyltransferase (AnPRT; trpD).
Evans, G.L., Baker, E.N., Lott, J.S.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Anthranilate phosphoribosyltransferase | 379 | Mycobacterium tuberculosis H37Ra | Mutation(s): 0 Gene Names: trpD, MRA_2208 EC: 2.4.2.18 | 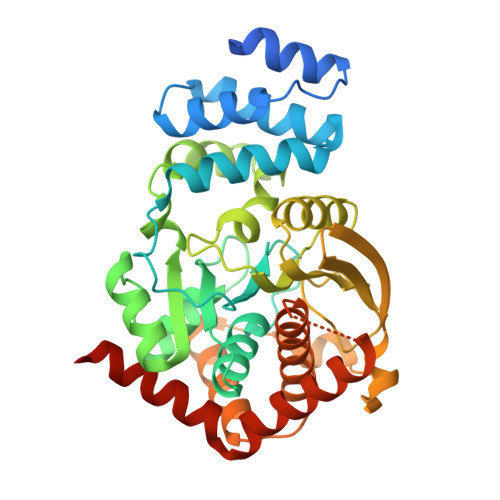 | |
UniProt | |||||
Find proteins for A5U4M0 (Mycobacterium tuberculosis (strain ATCC 25177 / H37Ra)) Explore A5U4M0 Go to UniProtKB: A5U4M0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5U4M0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PRP Query on PRP | E [auth A], J [auth B] | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose C5 H13 O14 P3 PQGCEDQWHSBAJP-TXICZTDVSA-N |  | ||
| 59L Query on 59L | F [auth A], K [auth B] | 2-[(2-carboxy-5-nitrophenyl)amino]-3-methylbenzoic acid C15 H12 N2 O6 LKOWRACJCMMHTL-UHFFFAOYSA-N |  | ||
| PO4 Query on PO4 | L [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| IMD Query on IMD | G [auth A] | IMIDAZOLE C3 H5 N2 RAXXELZNTBOGNW-UHFFFAOYSA-O |  | ||
| MG Query on MG | C [auth A], D [auth A], H [auth B], I [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 94.191 | α = 90 |
| b = 78.763 | β = 109.45 |
| c = 99.674 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data scaling |
| Aimless | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| Blu-Ice | data collection |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biopharma TRI Fund | New Zealand | 76673 |
| Health Research Council (HRC) | New Zealand | 12/110C |