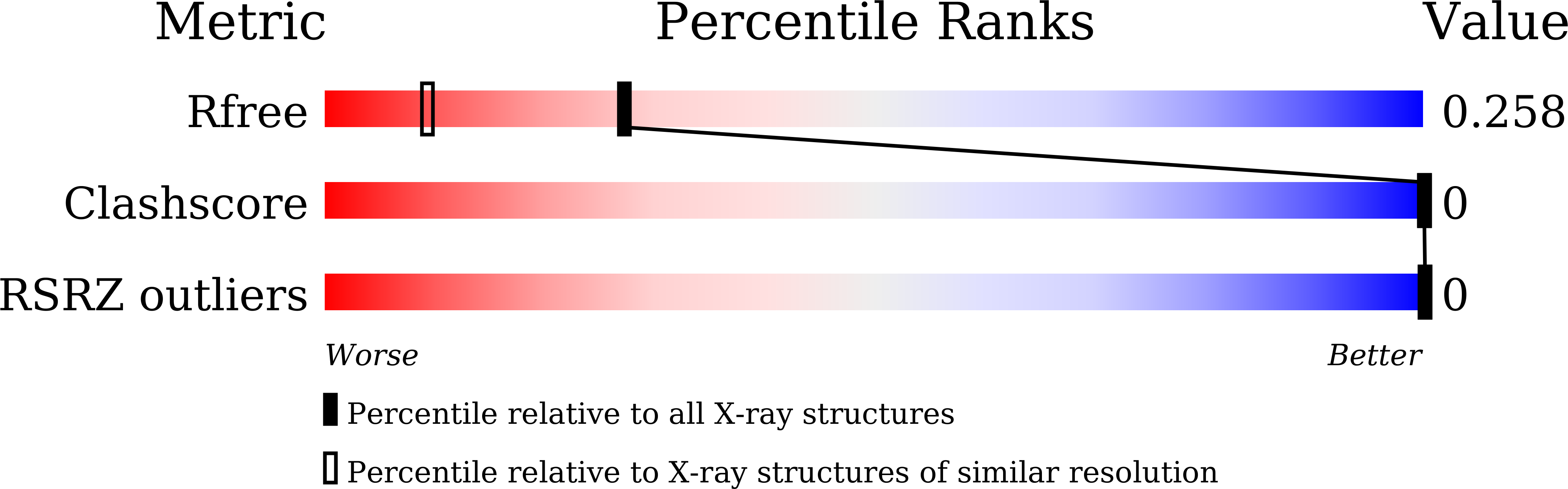The novel double-folded structure of d(GCATGCATGC): a possible model for triplet-repeat sequences
Thirugnanasambandam, A., Karthik, S., Mandal, P.K., Gautham, N.(2015) Acta Crystallogr D Biol Crystallogr 71: 2119-2126
- PubMed: 26457435
- DOI: https://doi.org/10.1107/S1399004715013930
- Primary Citation of Related Structures:
4ZKK - PubMed Abstract:
The structure of the decadeoxyribonucleotide d(GCATGCATGC) is presented at a resolution of 1.8 Å. The decamer adopts a novel double-folded structure in which the direction of progression of the backbone changes at the two thymine residues. Intra-strand stacking interactions (including an interaction between the endocylic O atom of a ribose moiety and the adjacent purine base), hydrogen bonds and cobalt-ion interactions stabilize the double-folded structure of the single strand. Two such double-folded strands come together in the crystal to form a dimer. Inter-strand Watson-Crick hydrogen bonds form four base pairs. This portion of the decamer structure is similar to that observed in other previously reported oligonucleotide structures and has been dubbed a `bi-loop'. Both the double-folded single-strand structure, as well as the dimeric bi-loop structure, serve as starting points to construct models for triplet-repeat DNA sequences, which have been implicated in many human diseases.
Organizational Affiliation:
CAS in Crystallography and Biophysics, University of Madras, Guindy Campus, Chennai, Tamil Nadu 600 025, India.