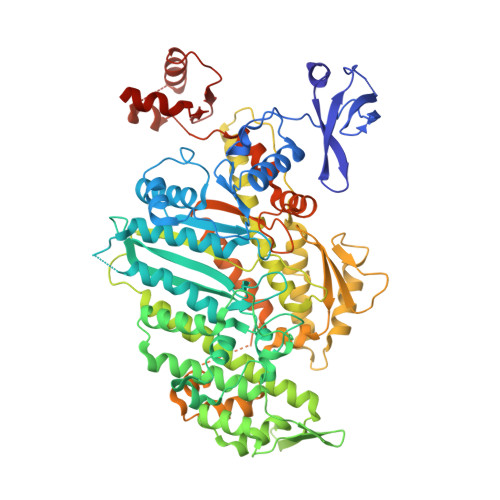
Force-producing ADP state of myosin bound to actin.
Wulf, S.F., Ropars, V., Fujita-Becker, S., Oster, M., Hofhaus, G., Trabuco, L.G., Pylypenko, O., Sweeney, H.L., Houdusse, A.M., Schroder, R.R.(2016) Proc Natl Acad Sci U S A 113: E1844-E1852
- PubMed: 26976594
- DOI: https://doi.org/10.1073/pnas.1516598113
- Primary Citation of Related Structures:
4ZG4 - PubMed Abstract:
Molecular motors produce force when they interact with their cellular tracks. For myosin motors, the primary force-generating state has MgADP tightly bound, whereas myosin is strongly bound to actin. We have generated an 8-Å cryoEM reconstruction of this state for myosin V and used molecular dynamics flexed fitting for model building. We compare this state to the subsequent state on actin (Rigor). The ADP-bound structure reveals that the actin-binding cleft is closed, even though MgADP is tightly bound. This state is accomplished by a previously unseen conformation of the β-sheet underlying the nucleotide pocket. The transition from the force-generating ADP state to Rigor requires a 9.5° rotation of the myosin lever arm, coupled to a β-sheet rearrangement. Thus, the structure reveals the detailed rearrangements underlying myosin force generation as well as the basis of strain-dependent ADP release that is essential for processive myosins, such as myosin V.
Organizational Affiliation:
Cryo Electron Microscopy, CellNetworks, BioQuant, Universitätsklinikum Heidelberg, 69120 Heidelberg, Germany;