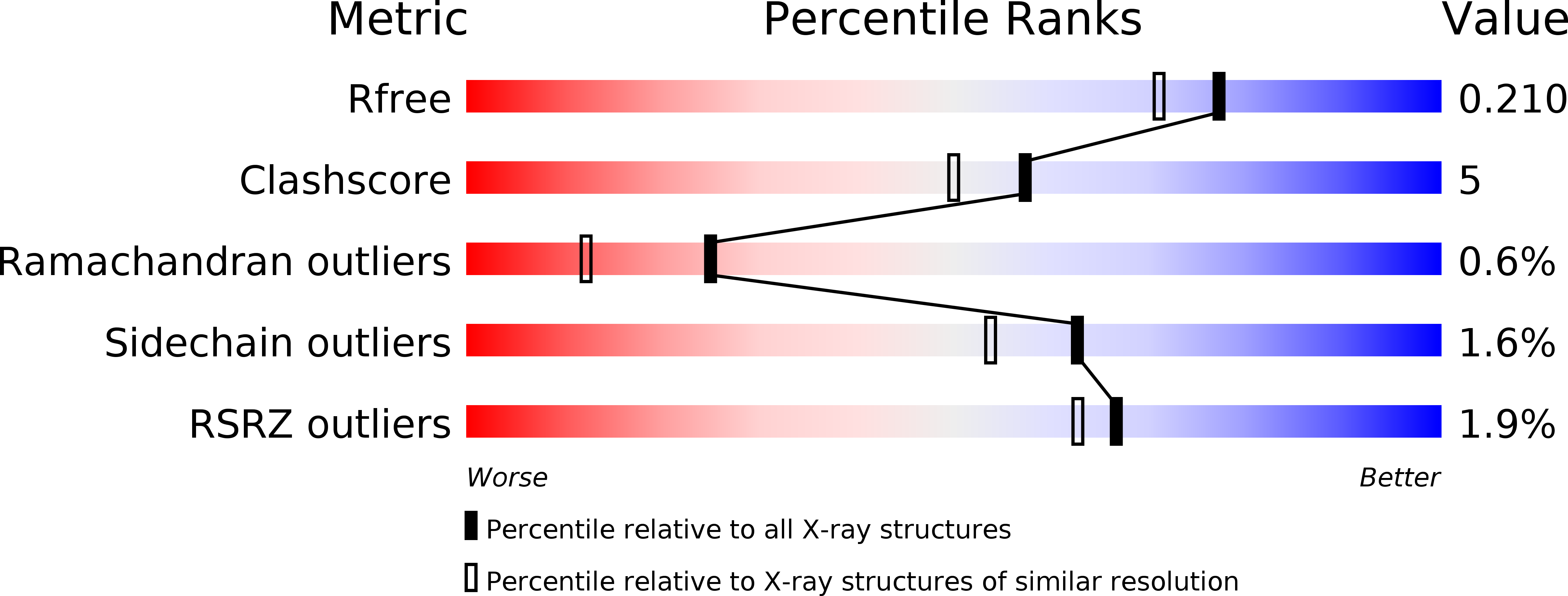
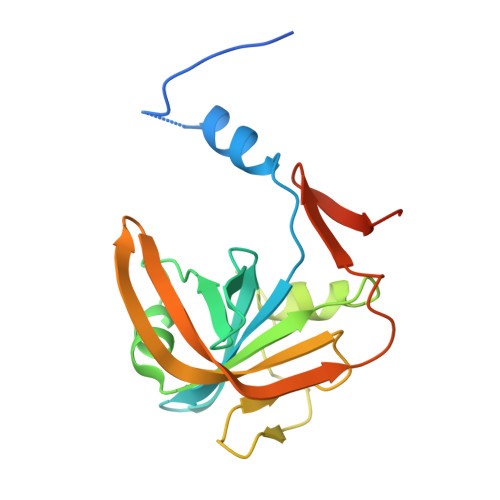
Biochemical properties and crystal structure of the flavin reductase FerA from Paracoccus denitrificans.
Sedlacek, V., Klumpler, T., Marek, J., Kucera, I.(2016) Microbiol Res 188-189: 9-22
- PubMed: 27296958
- DOI: https://doi.org/10.1016/j.micres.2016.04.006
- Primary Citation of Related Structures:
4XHY, 4XJ2 - PubMed Abstract:
The Pden_2689 gene encoding FerA, an NADH:flavin oxidoreductase required for growth of Paracoccus denitrificans under iron limitation, was cloned and overexpressed as a C-terminally His6-tagged derivative. The binding of substrates and products was detected and quantified by isothermal titration calorimetry and fluorometric titration. FerA binds FMN and FAD with comparable affinity in an enthalpically driven, entropically opposed process. The reduced flavin is bound more loosely than the oxidized one, which was confirmed by a negative shift in the redox potential of FMN after addition of FerA. Initial velocity and substrate analogs inhibition studies showed that FerA follows a random-ordered sequence of substrate (NADH and FMN) binding. The primary kinetic isotope effects from stereospecifically deuterated nicotinamide nucleotides demonstrated that hydride transfer occurs from the pro-S position and contributes to rate limitation for the overall reaction. The crystal structure of FerA revealed a twisted seven-stranded antiparallel β-barrel similar to that of other short chain flavin reductases. Only minor structural changes around Arg106 took place upon FMN binding. The solution structure FerA derived from small angle X-ray scattering (SAXS) matched the dimer assembly predicted from the crystal structure. Site-directed mutagenesis pinpointed a role of Arg106 and His146 in binding of flavin and NADH, respectively. Pull down experiments performed with cytoplasmic extracts resulted in a negative outcome indicating that FerA might physiologically act without association with other proteins. Rapid kinetics experiments provided evidence for a stabilizing effect of another P. denitrificans protein, the
Organizational Affiliation:
Department of Biochemistry, Faculty of Science, Masaryk University, Brno, Czech Republic.