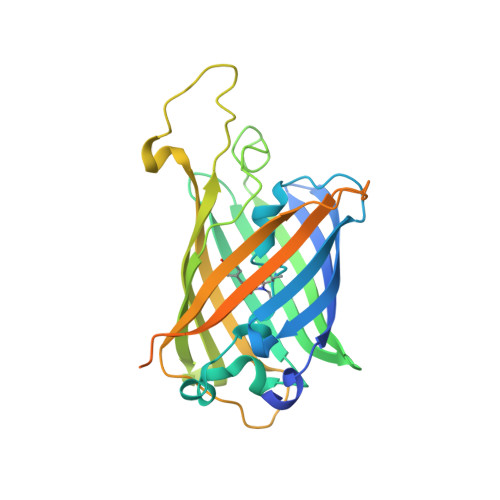
The structure of a GFP-based antibody (fluorobody) to TLH, a toxin from Vibrio parahaemolyticus.
Chen, Y., Huang, X., Wang, R., Wang, S., Shi, N.(2015) Acta Crystallogr F Struct Biol Commun 71: 913-918
- PubMed: 26144238
- DOI: https://doi.org/10.1107/S2053230X15008845
- Primary Citation of Related Structures:
4XGY - PubMed Abstract:
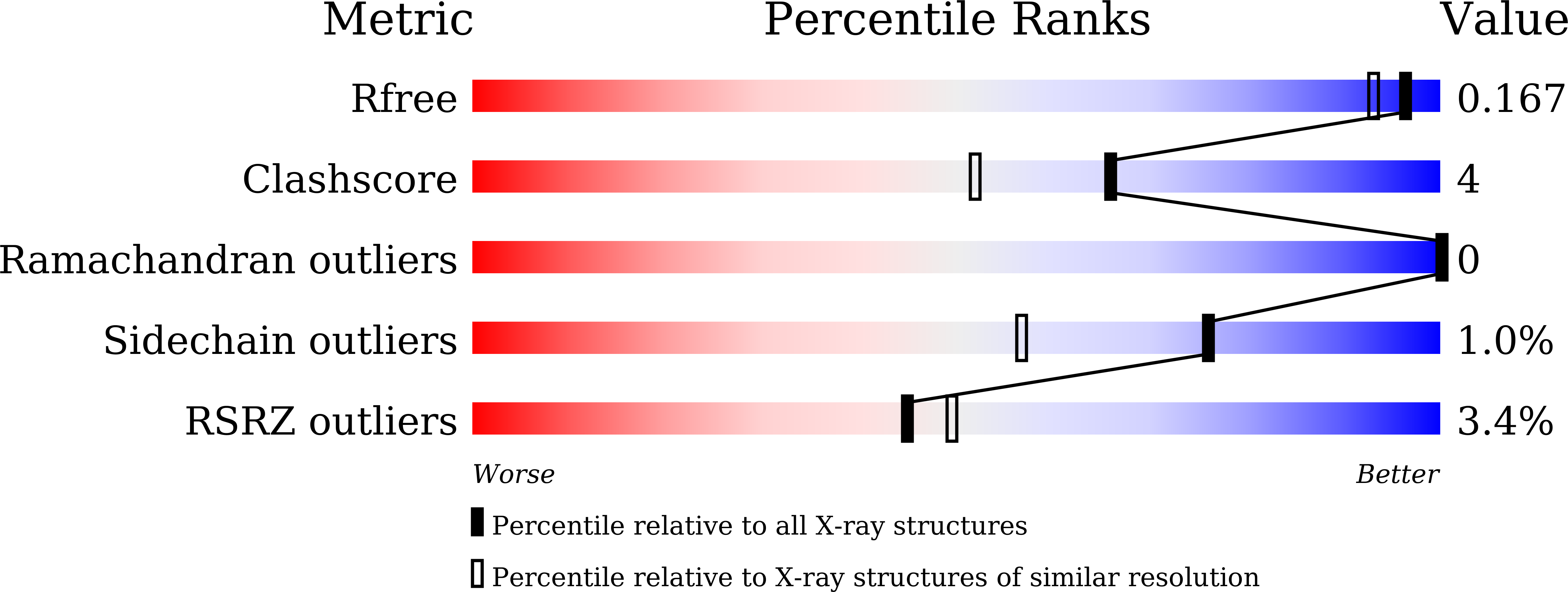
A fluorobody is a manmade hybrid molecule that is composed of green fluorescent protein (GFP) and a fragment of antibody, which combines the affinity and specificity of an antibody with the visibility of a GFP. It is able to provide a real-time indication of binding while avoiding the use of tags and secondary binding reagents. Here, the expression, purification and crystal structure of a recombinant fluorobody for TLH (thermolabile haemolysin), a toxin from the lethal food-borne disease bacterium Vibrio parahaemolyticus, are presented. This is the first structure of a fluorobody to be reported. Crystals belonging to space group P4(3)2(1)2, with unit-cell parameters a = b = 63.35, c = 125.90 Å, were obtained by vapour diffusion in hanging drops and the structure was refined to an Rfree of 16.7% at 1.5 Å resolution. The structure shows a CDR loop of the antibody on the GFP scaffold.
Organizational Affiliation:
State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, 155 Yangqiao Road West, Fuzhou 350002, People's Republic of China.