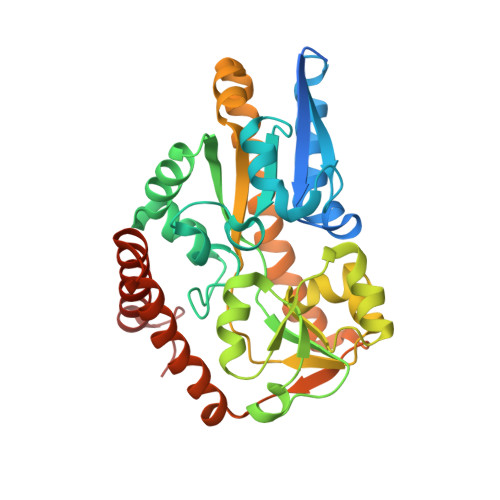
Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound (S)-(+)-2-Amino-1-propanol.
Vetting, M.W., Al Obaidi, N.F., Toro, R., Morisco, L.L., Benach, J., Wasserman, S.R., Attonito, J.D., Scott Glenn, A., Chamala, S., Chowdhury, S., Lafleur, J., Love, J., Seidel, R.D., Whalen, K.L., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.