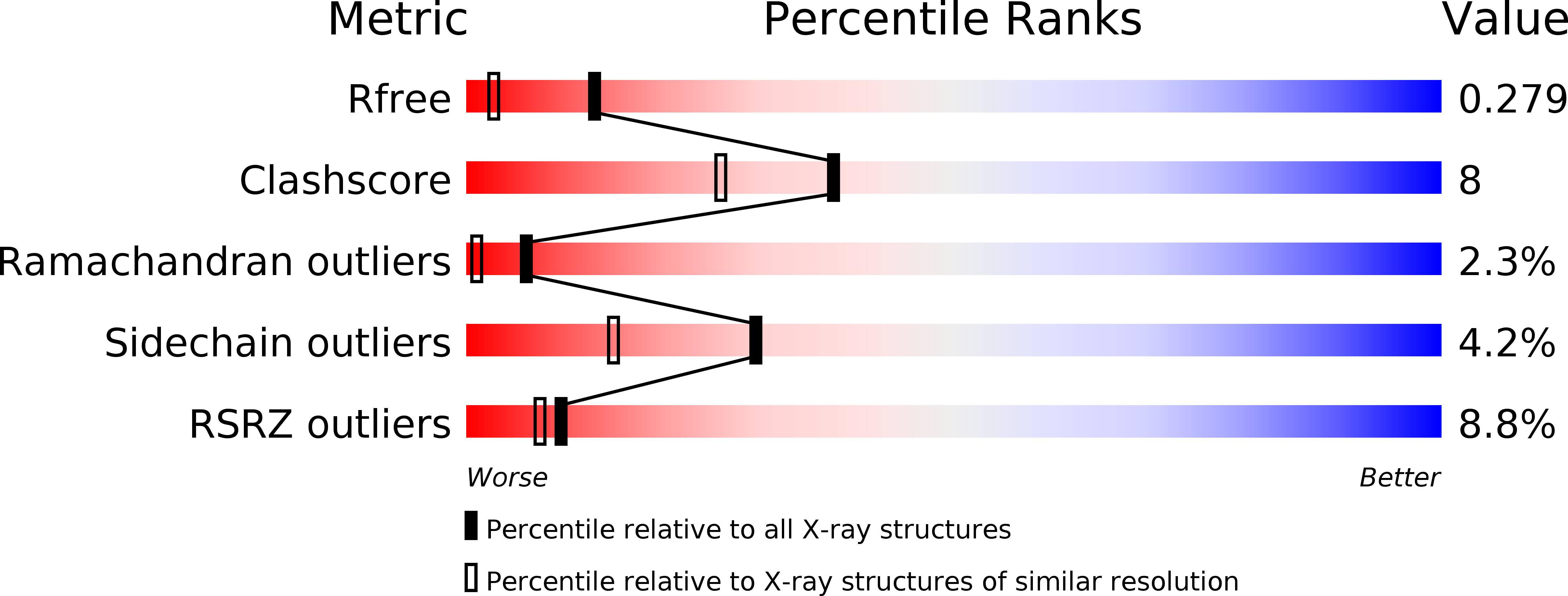
PspF-binding domain PspA1-144 and the PspAF complex: New insights into the coiled-coil-dependent regulation of AAA+ proteins.
Osadnik, H., Schopfel, M., Heidrich, E., Mehner, D., Lilie, H., Parthier, C., Risselada, H.J., Grubmuller, H., Stubbs, M.T., Bruser, T.(2015) Mol Microbiol 98: 743-759
- PubMed: 26235546
- DOI: https://doi.org/10.1111/mmi.13154
- Primary Citation of Related Structures:
4WHE - PubMed Abstract:
Phage shock protein A (PspA) belongs to the highy conserved PspA/IM30 family and is a key component of the stress inducible Psp system in Escherichia coli. One of its central roles is the regulatory interaction with the transcriptional activator of this system, the σ(54) enhancer-binding protein PspF, a member of the AAA+ protein family. The PspA/F regulatory system has been intensively studied and serves as a paradigm for AAA+ enzyme regulation by trans-acting factors. However, the molecular mechanism of how exactly PspA controls the activity of PspF and hence σ(54) -dependent expression of the psp genes is still unclear. To approach this question, we identified the minimal PspF-interacting domain of PspA, solved its structure, determined its affinity to PspF and the dissociation kinetics, identified residues that are potentially important for PspF regulation and analyzed effects of their mutation on PspF in vivo and in vitro. Our data indicate that several characteristics of AAA+ regulation in the PspA·F complex resemble those of the AAA+ unfoldase ClpB, with both proteins being regulated by a structurally highly conserved coiled-coil domain. The convergent evolution of both regulatory domains points to a general mechanism to control AAA+ activity for divergent physiologic tasks via coiled-coil domains.
Organizational Affiliation:
Institute of Microbiology, Leibniz Universität Hannover, Herrenhäuser Str. 2, Hannover, 30419, Germany.