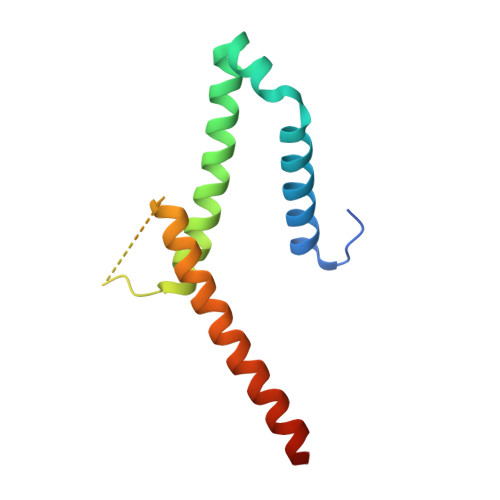
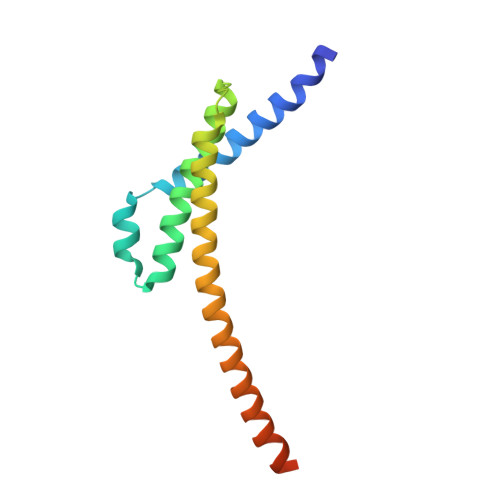
The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Schuch, B., Feigenbutz, M., Makino, D.L., Falk, S., Basquin, C., Mitchell, P., Conti, E.(2014) EMBO J 33: 2829-2846
- PubMed: 25319414
- DOI: https://doi.org/10.15252/embj.201488757
- Primary Citation of Related Structures:
4WFC, 4WFD - PubMed Abstract:
The exosome is a conserved multi-subunit ribonuclease complex that functions in 3' end processing, turnover and surveillance of nuclear and cytoplasmic RNAs. In the yeast nucleus, the 10-subunit core complex of the exosome (Exo-10) physically and functionally interacts with the Rrp6 exoribonuclease and its associated cofactor Rrp47, the helicase Mtr4 and Mpp6. Here, we show that binding of Mtr4 to Exo-10 in vitro is dependent upon both Rrp6 and Rrp47, whereas Mpp6 binds directly and independently of other cofactors. Crystallographic analyses reveal that the N-terminal domains of Rrp6 and Rrp47 form a highly intertwined structural unit. Rrp6 and Rrp47 synergize to create a composite and conserved surface groove that binds the N-terminus of Mtr4. Mutation of conserved residues within Rrp6 and Mtr4 at the structural interface disrupts their interaction and inhibits growth of strains expressing a C-terminal GFP fusion of Mtr4. These studies provide detailed structural insight into the interaction between the Rrp6-Rrp47 complex and Mtr4, revealing an important link between Mtr4 and the core exosome.
Organizational Affiliation:
Structural Cell Biology Department, Max Planck Institute of Biochemistry, Martinsried, Germany.