Mechanism of Thiosulfate Oxidation in the Soxa Family of Cysteine-Ligated Cytochromes
Grabarczyk, D.B., Chappell, P.E., Eisel, B., Johnson, S., Lea, S.M., Berks, B.C.(2015) J Biol Chem 290: 9209
- PubMed: 25673696
- DOI: https://doi.org/10.1074/jbc.M114.618025
- Primary Citation of Related Structures:
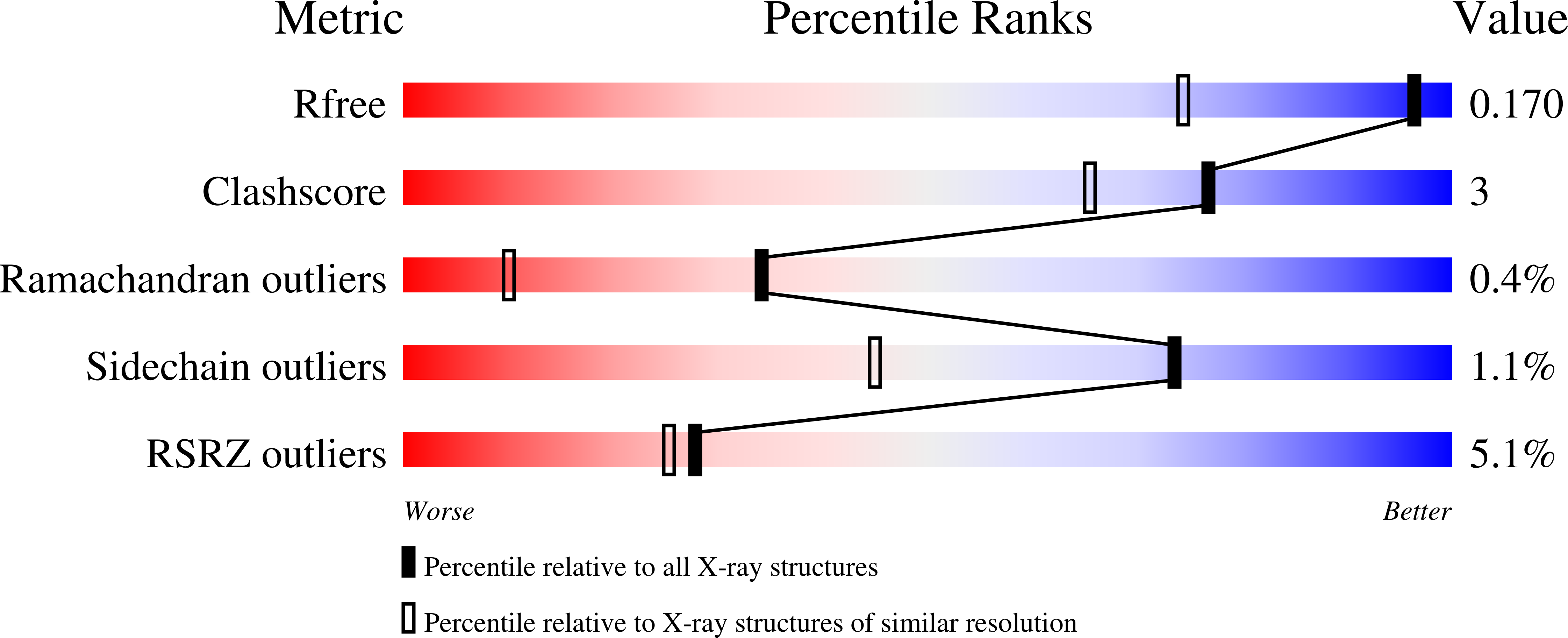
4V2K - PubMed Abstract:
Thiosulfate dehydrogenase (TsdA) catalyzes the oxidation of two thiosulfate molecules to form tetrathionate and is predicted to use an unusual cysteine-ligated heme as the catalytic cofactor. We have determined the structure of Allochromatium vinosum TsdA to a resolution of 1.3 Å. This structure confirms the active site heme ligation, identifies a thiosulfate binding site within the active site cavity, and reveals an electron transfer route from the catalytic heme, through a second heme group to the external electron acceptor. We provide multiple lines of evidence that the catalytic reaction proceeds through the intermediate formation of a S-thiosulfonate derivative of the heme cysteine ligand: the cysteine is reactive and is accessible to electrophilic attack; cysteine S-thiosulfonate is formed by the addition of thiosulfate or following the reverse reaction with tetrathionate; the S-thiosulfonate modification is removed through catalysis; and alkylating the cysteine blocks activity. Active site amino acid residues required for catalysis were identified by mutagenesis and are inferred to also play a role in stabilizing the S-thiosulfonate intermediate. The enzyme SoxAX, which catalyzes the first step in the bacterial Sox thiosulfate oxidation pathway, is homologous to TsdA and can be inferred to use a related catalytic mechanism.
Organizational Affiliation:
From the Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, United Kingdom and.