Skelemin Association with Alphaiibbeta3 Integrin: A Structural Model.
Gorbatyuk, V., Nguyen, K., Podolnikova, N.P., Deshmukh, L., Lin, X., Ugarova, T.P., Vinogradova, O.(2014) Biochemistry 53: 6766-6775
- PubMed: 25224262
- DOI: https://doi.org/10.1021/bi500680s
- Primary Citation of Related Structures:
4V10 - PubMed Abstract:
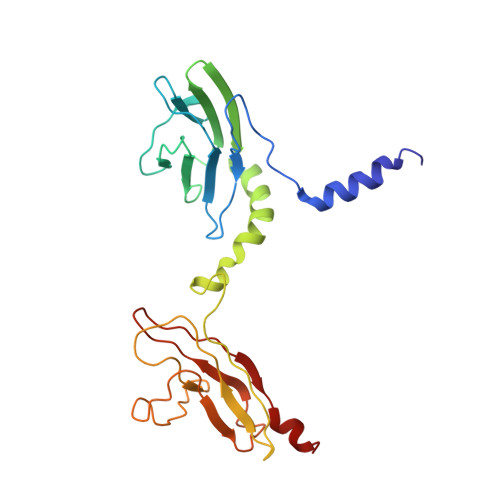
Over the last two decades, our knowledge concerning intracellular events that regulate integrin's affinity to their soluble ligands has significantly improved. However, the mechanism of adhesion-induced integrin clustering and development of focal complexes, which could further mature to form focal adhesions, still remains under-investigated. Here we present a structural model of tandem IgC2 domains of skelemin in complex with the cytoplasmic tails of integrin αIIbβ3. The model of tertiary assembly is generated based upon NMR data and illuminates a potential link between the essential cell adhesion receptors and myosin filaments. This connection may serve as a basis for generating the mechanical forces necessary for cell migration and remodeling.
Organizational Affiliation:
Department of Pharmaceutical Sciences, School of Pharmacy, University of Connecticut at Storrs , Storrs, Connecticut 06269, United States.