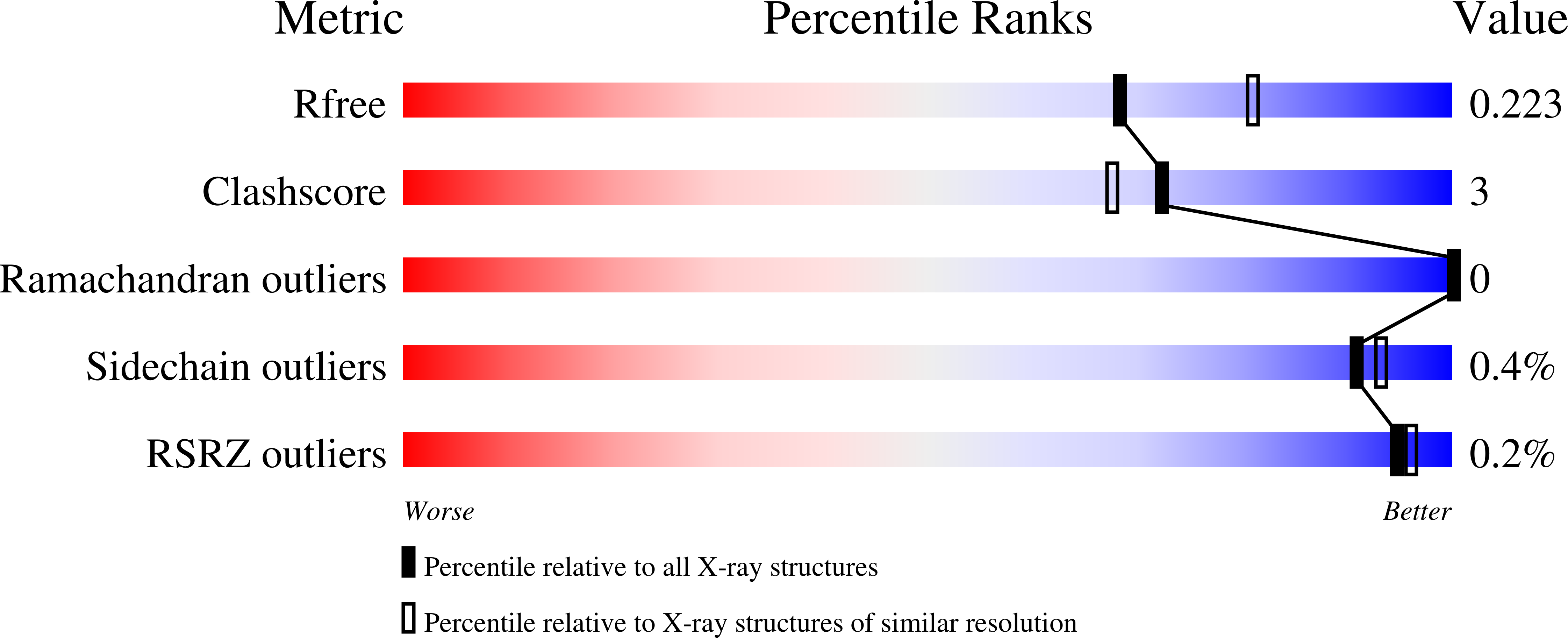
13-methylarachidonic Acid is a positive allosteric modulator of endocannabinoid oxygenation by cyclooxygenase.
Kudalkar, S.N., Nikas, S.P., Kingsley, P.J., Xu, S., Galligan, J.J., Rouzer, C.A., Banerjee, S., Ji, L., Eno, M.R., Makriyannis, A., Marnett, L.J.(2015) J Biol Chem 290: 7897-7909
- PubMed: 25648895
- DOI: https://doi.org/10.1074/jbc.M114.634014
- Primary Citation of Related Structures:
4RUT - PubMed Abstract:
Cyclooxygenase-2 (COX-2) oxygenates arachidonic acid (AA) and the endocannabinoids 2-arachidonoylglycerol (2-AG) and arachidonylethanolamide to prostaglandins, prostaglandin glyceryl esters, and prostaglandin ethanolamides, respectively. A structural homodimer, COX-2 acts as a conformational heterodimer with a catalytic and an allosteric monomer. Prior studies have demonstrated substrate-selective negative allosteric regulation of 2-AG oxygenation. Here we describe AM-8138 (13(S)-methylarachidonic acid), a substrate-selective allosteric potentiator that augments 2-AG oxygenation by up to 3.5-fold with no effect on AA oxygenation. In the crystal structure of an AM-8138·COX-2 complex, AM-8138 adopts a conformation similar to the unproductive conformation of AA in the substrate binding site. Kinetic analysis suggests that binding of AM-8138 to the allosteric monomer of COX-2 increases 2-AG oxygenation by increasing kcat and preventing inhibitory binding of 2-AG. AM-8138 restored the activity of COX-2 mutants that exhibited very poor 2-AG oxygenating activity and increased the activity of COX-1 toward 2-AG. Competition of AM-8138 for the allosteric site prevented the inhibition of COX-2-dependent 2-AG oxygenation by substrate-selective inhibitors and blocked the inhibition of AA or 2-AG oxygenation by nonselective time-dependent inhibitors. AM-8138 selectively enhanced 2-AG oxygenation in intact RAW264.7 macrophage-like cells. Thus, AM-8138 is an important new tool compound for the exploration of allosteric modulation of COX enzymes and their role in endocannabinoid metabolism.
Organizational Affiliation:
From the A. B. Hancock Jr. Memorial Laboratory for Cancer Research, Departments of Biochemistry.