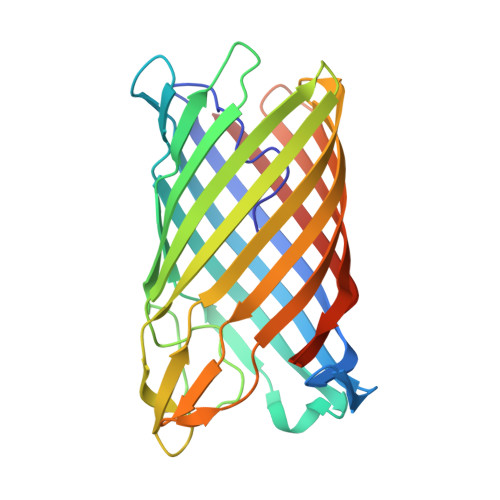
Crystal structure of a COG4313 outer membrane channel.
Berg, B.v., Bhamidimarri, S.P., Winterhalter, M.(2015) Sci Rep 5: 11927-11927
- PubMed: 26149193
- DOI: https://doi.org/10.1038/srep11927
- Primary Citation of Related Structures:
4RL8 - PubMed Abstract:
COG4313 proteins form a large and widespread family of outer membrane channels and have been implicated in the uptake of a variety of hydrophobic molecules. Structure-function studies of this protein family have so far been hampered by a lack of structural information. Here we present the X-ray crystal structure of Pput2725 from the biodegrader Pseudomonas putida F1, a COG4313 channel of unknown function, using data to 2.3 Å resolution. The structure shows a 12-stranded barrel with an N-terminal segment preceding the first β-strand occluding the lumen of the barrel. Single channel electrophysiology and liposome swelling experiments suggest that while the narrow channel visible in the crystal structure does allow passage of ions and certain small molecules in vitro, Pput2725 is unlikely to function as a channel for hydrophilic molecules. Instead, the presence of bound detergent molecules inside the barrel suggests that Pput2725 mediates uptake of hydrophobic molecules. Sequence alignments and the locations of highly conserved residues suggest the presence of a dynamic lateral opening through which hydrophobic molecules might gain entry into the cell. Our results provide the basis for structure-function studies of COG4313 family members with known function, such as the SphA sphingosine uptake channel of Pseudomonas aeruginosa.
Organizational Affiliation:
Institute for Cell and Molecular Biosciences, The Medical School, Newcastle University, Newcastle upon Tyne, NE2 4HH, UK.