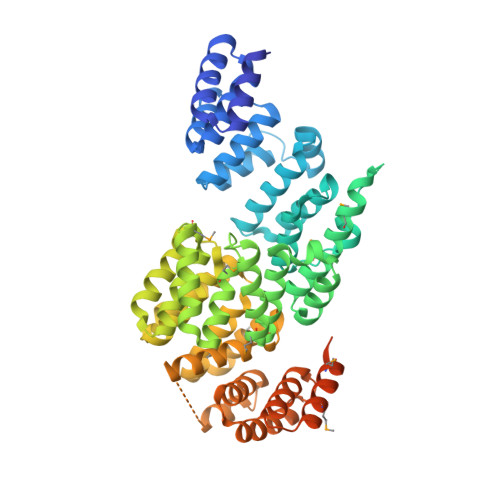
Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
Yamaguchi, M., Yu, S., Qiao, R., Weissmann, F., Miller, D.J., VanderLinden, R., Brown, N.G., Frye, J.J., Peters, J.M., Schulman, B.A.(2015) J Mol Biol 427: 1748-1764
- PubMed: 25490258
- DOI: https://doi.org/10.1016/j.jmb.2014.11.020
- Primary Citation of Related Structures:
4RG6, 4RG7, 4RG9 - PubMed Abstract:
The anaphase-promoting complex/cyclosome (APC/C) is a massive E3 ligase that controls mitosis by catalyzing ubiquitination of key cell cycle regulatory proteins. The APC/C assembly contains two subcomplexes: the "Platform" centers around a cullin-RING-like E3 ligase catalytic core; the "Arc Lamp" is a hub that mediates transient association with regulators and ubiquitination substrates. The Arc Lamp contains the small subunits APC16, CDC26, and APC13, and tetratricopeptide repeat (TPR) proteins (APC7, APC3, APC6, and APC8) that homodimerize and stack with quasi-2-fold symmetry. Within the APC/C complex, APC3 serves as center for regulation. APC3's TPR motifs recruit substrate-binding coactivators, CDC20 and CDH1, via their C-terminal conserved Ile-Arg (IR) tail sequences. Human APC3 also binds APC16 and APC7 and contains a >200-residue loop that is heavily phosphorylated during mitosis, although the basis for APC3 interactions and whether loop phosphorylation is required for ubiquitination are unclear. Here, we map the basis for human APC3 assembly with APC16 and APC7, report crystal structures of APC3Δloop alone and in complex with the C-terminal domain of APC16, and test roles of APC3's loop and IR tail binding surfaces in APC/C-catalyzed ubiquitination. The structures show how one APC16 binds asymmetrically to the symmetric APC3 dimer and, together with biochemistry and prior data, explain how APC16 recruits APC7 to APC3, show how APC3's C-terminal domain is rearranged in the full APC/C assembly, and visualize residues in the IR tail binding cleft important for coactivator-dependent ubiquitination. Overall, the results provide insights into assembly, regulation, and interactions of TPR proteins and the APC/C.
Organizational Affiliation:
Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN 38105, USA.