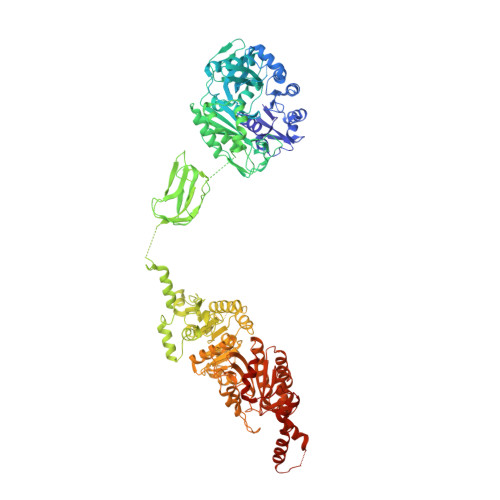
Structure and function of a single-chain, multi-domain long-chain acyl-CoA carboxylase.
Tran, T.H., Hsiao, Y.S., Jo, J., Chou, C.Y., Dietrich, L.E., Walz, T., Tong, L.(2015) Nature 518: 120-124
- PubMed: 25383525
- DOI: https://doi.org/10.1038/nature13912
- Primary Citation of Related Structures:
4RCN - PubMed Abstract:
Biotin-dependent carboxylases are widely distributed in nature and have important functions in the metabolism of fatty acids, amino acids, carbohydrates, cholesterol and other compounds. Defective mutations in several of these enzymes have been linked to serious metabolic diseases in humans, and acetyl-CoA carboxylase is a target for drug discovery in the treatment of diabetes, cancer and other diseases. Here we report the identification and biochemical, structural and functional characterizations of a novel single-chain (120 kDa), multi-domain biotin-dependent carboxylase in bacteria. It has preference for long-chain acyl-CoA substrates, although it is also active towards short-chain and medium-chain acyl-CoAs, and we have named it long-chain acyl-CoA carboxylase. The holoenzyme is a homo-hexamer with molecular mass of 720 kDa. The 3.0 Å crystal structure of the long-chain acyl-CoA carboxylase holoenzyme from Mycobacterium avium subspecies paratuberculosis revealed an architecture that is strikingly different from those of related biotin-dependent carboxylases. In addition, the domains of each monomer have no direct contact with each other. They are instead extensively swapped in the holoenzyme, such that one cycle of catalysis involves the participation of four monomers. Functional studies in Pseudomonas aeruginosa suggest that the enzyme is involved in the utilization of selected carbon and nitrogen sources.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York 10027, USA.