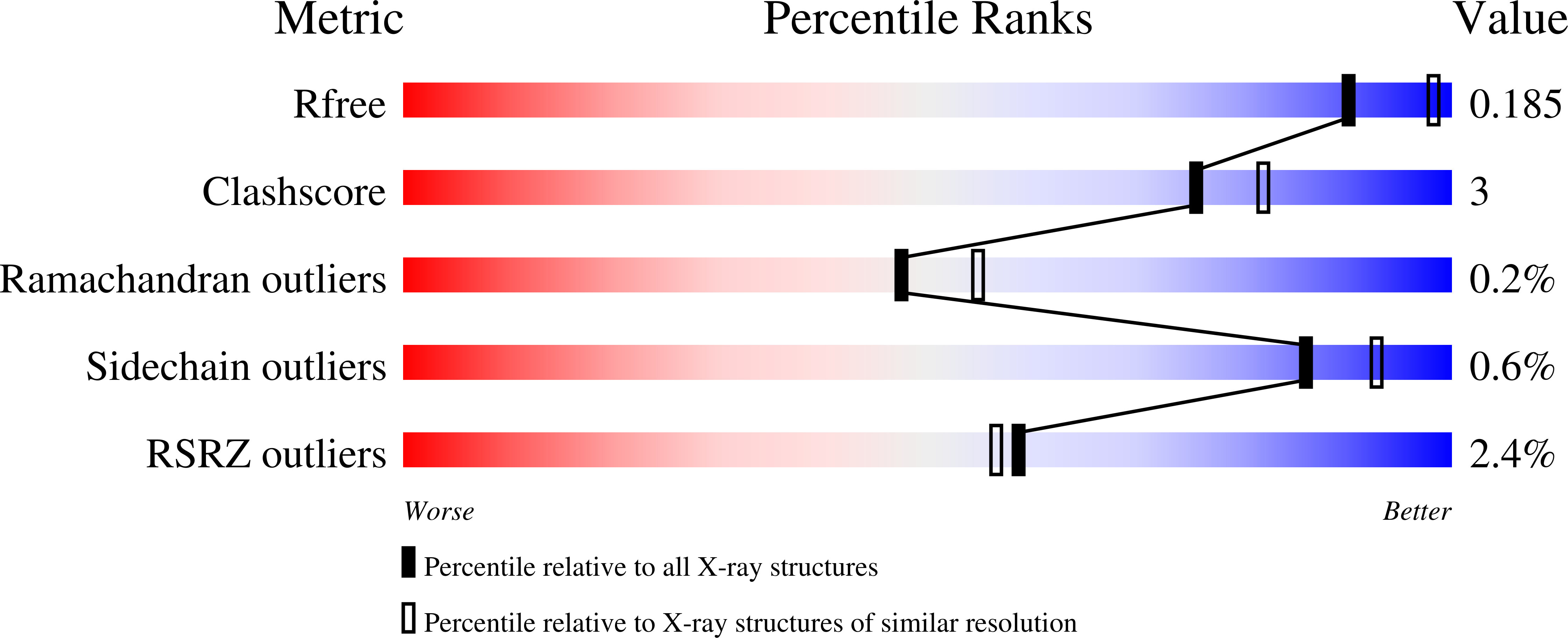
The kinetic characterization and X-ray structure of a putative benzoylformate decarboxylase from M. smegmatis highlights the difficulties in the functional annotation of ThDP-dependent enzymes.
Andrews, F.H., Horton, J.D., Shin, D., Yoon, H.J., Logsdon, M.G., Malik, A.M., Rogers, M.P., Kneen, M.M., Suh, S.W., McLeish, M.J.(2015) Biochim Biophys Acta 1854: 1001-1009
- PubMed: 25936776
- DOI: https://doi.org/10.1016/j.bbapap.2015.04.027
- Primary Citation of Related Structures:
4Q9D - PubMed Abstract:
Benzoylformate decarboxylase (BFDC) is a thiamin diphosphate (ThDP)-dependent enzyme that catalyzes the nonoxidative decarboxylation of benzoylformate. It is the penultimate enzyme in both the mandelate pathway and the d-phenylglycine degradation pathway. The ThDP-dependent Enzyme Engineering Database (TEED) now lists more than 800 sequences annotated as BFDCs, including one from Mycobacterium smegmatis (MsBFDC). However, there is no evidence that either pathway for benzoylformate formation exists in the M. smegmatis genome. Further, sequence alignments of MsBFDC with the well characterized enzyme isolated from Pseudomonas putida (PpBFDC) indicate that there will be active site substitutions in MsBFDC likely to reduce activity with benzoylformate. Taken together these data would suggest that the annotation is unlikely to be correct. To test this hypothesis the putative MsBFDC was cloned, expressed, purified, and the X-ray structure was solved to a resolution of 2.2Å. While showing no evidence for ThDP in the active site, the structure was very similar to that of PpBFDC. A number of 2-oxo acids were tested as substrates. For MsBFDC the K(m) value for benzoylformate was ~23 mM, nearly 100-fold greater than that of PpBFDC while the k(cat) value was reduced 60-fold. These values would suggest that benzoylformate is not the physiological substrate for this enzyme, and that annotation as a 2-oxo acid decarboxylase may be more appropriate.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Indiana University-Purdue University Indianapolis (IUPUI), Indianapolis, IN 46202, USA.