Functional and structural properties of a novel protein and virulence factor (Protein sHIP) in Streptococcus pyogenes.
Wisniewska, M., Happonen, L., Kahn, F., Varjosalo, M., Malmstrom, L., Rosenberger, G., Karlsson, C., Cazzamali, G., Pozdnyakova, I., Frick, I.M., Bjorck, L., Streicher, W., Malmstrom, J., Wikstrom, M.(2014) J Biol Chem 289: 18175-18188
- PubMed: 24825900
- DOI: https://doi.org/10.1074/jbc.M114.565978
- Primary Citation of Related Structures:
4MER, 4PZ1 - PubMed Abstract:
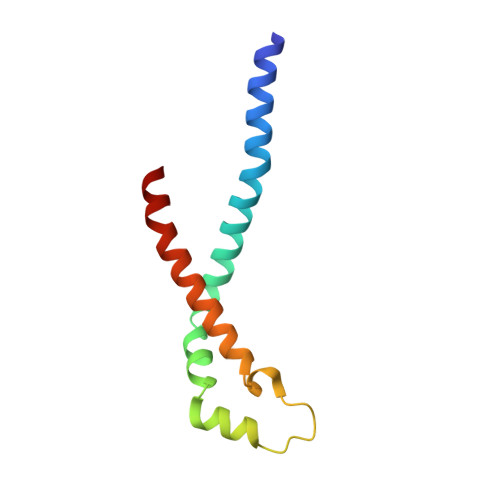
Streptococcus pyogenes is a significant bacterial pathogen in the human population. The importance of virulence factors for the survival and colonization of S. pyogenes is well established, and many of these factors are exposed to the extracellular environment, enabling bacterial interactions with the host. In the present study, we quantitatively analyzed and compared S. pyogenes proteins in the growth medium of a strain that is virulent to mice with a non-virulent strain. Particularly, one of these proteins was present at significantly higher levels in stationary growth medium from the virulent strain. We determined the three-dimensional structure of the protein that showed a unique tetrameric organization composed of four helix-loop-helix motifs. Affinity pull-down mass spectrometry analysis in human plasma demonstrated that the protein interacts with histidine-rich glycoprotein (HRG), and the name sHIP (streptococcal histidine-rich glycoprotein-interacting protein) is therefore proposed. HRG has antibacterial activity, and when challenged by HRG, sHIP was found to rescue S. pyogenes bacteria. This and the finding that patients with invasive S. pyogenes infection respond with antibody production against sHIP suggest a role for the protein in S. pyogenes pathogenesis.
Organizational Affiliation:
From the Novo Nordisk Foundation Center for Protein Research, University of Copenhagen, DK-2200 Copenhagen, Denmark.