Crystal structure of CbXyn10B from Caldicellulosiruptor bescii and its mutant(E139A) in complex with hydrolyzed xylotetraose
An, J., Wu, G., Feng, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Endo-1,4-beta-xylanase | 338 | Caldicellulosiruptor bescii DSM 6725 | Mutation(s): 1 Gene Names: Athe_0185 EC: 3.2.1.8 | 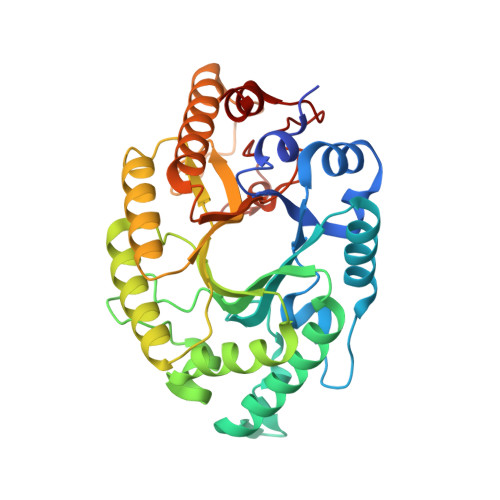 | |
UniProt | |||||
Find proteins for B9MMA5 (Caldicellulosiruptor bescii (strain ATCC BAA-1888 / DSM 6725 / KCTC 15123 / Z-1320)) Explore B9MMA5 Go to UniProtKB: B9MMA5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B9MMA5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900116 Query on PRD_900116 | B | 4beta-beta-xylobiose | Oligosaccharide / Metabolism |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.319 | α = 90 |
| b = 80.817 | β = 90 |
| c = 115.19 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data reduction |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL | data scaling |