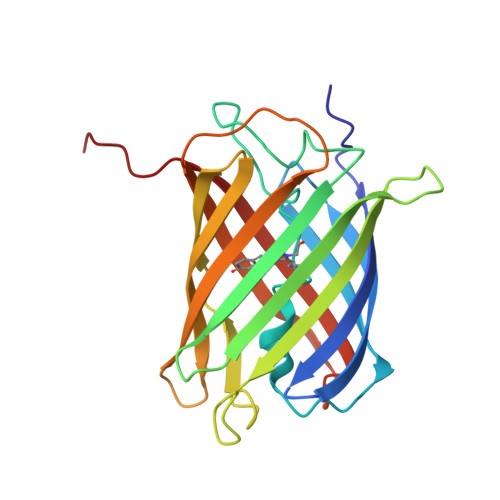
A diffraction-quality protein crystal processed as an autophagic cargo
Tsutsui, H., Jinno, Y., Shoda, K., Tomita, A., Matsuda, M., Yamashita, E., Katayama, H., Nakagawa, A., Miyawaki, A.(2015) Mol Cell 58: 186-193
- PubMed: 25773597
- DOI: https://doi.org/10.1016/j.molcel.2015.02.007
- Primary Citation of Related Structures:
4P76 - PubMed Abstract:
Crystallization of proteins may occur in the cytosol of a living cell, but how a cell responds to intracellular protein crystallization remains unknown. We developed a variant of coral fluorescent protein that forms diffraction-quality crystals within mammalian cells. This expression system allowed the direct determination of its crystal structure at 2.9 Å, as well as observation of the crystallization process and cellular responses. The micron-sized crystal, which emerged rapidly, was a pure assembly of properly folded β-barrels and was recognized as an autophagic cargo that was transferred to lysosomes via a process involving p62 and LC3. Several lines of evidence indicated that autophagy was not required for crystal nucleation or growth. These findings demonstrate that in vivo protein crystals can provide an experimental model to study chemical catalysis. This knowledge may be beneficial for structural biology studies on normal and disease-related protein aggregation.
Organizational Affiliation:
Laboratory for Cell Function Dynamics, Brain Science Institute, RIKEN, Wako, Saitama 351-0198, Japan; Department of Material Science, Japan Advanced Institute of Science and Technology, Nomi, Ishikawa 923-1292, Japan; PRESTO, Japan Science and Technology Agency (JST), 4-1-8 Hon-cho, Kawaguchi, Saitama 351-0198, Japan.