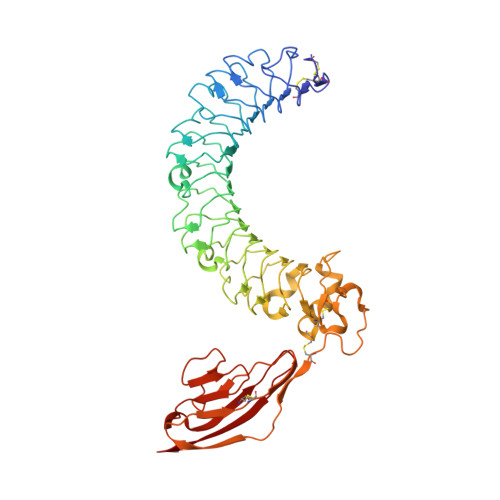
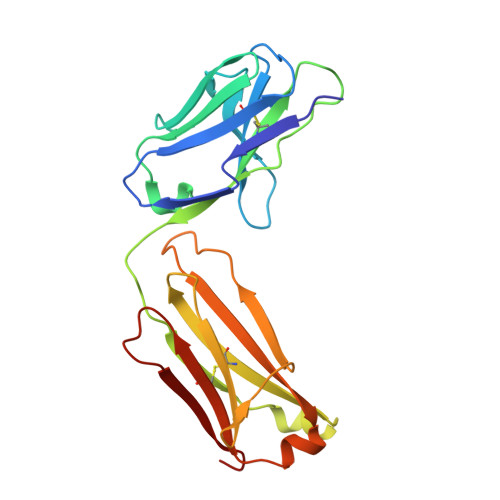
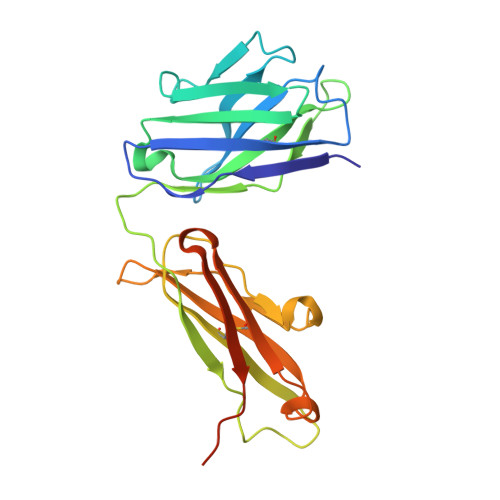
Structure of the LINGO-1-Anti-LINGO-1 Li81 Antibody Complex Provides Insights into the Biology of LINGO-1 and the Mechanism of Action of the Antibody Therapy.
Pepinsky, R.B., Arndt, J.W., Quan, C., Gao, Y., Quintero-Monzon, O., Lee, X., Mi, S.(2014) J Pharmacol Exp Ther 350: 110-123
- PubMed: 24756303
- DOI: https://doi.org/10.1124/jpet.113.211771
- Primary Citation of Related Structures:
4OQT - PubMed Abstract:
Multiple sclerosis (MS) is an autoimmune-inflammatory disease of the central nervous system (CNS) with prominent demyelination and axonal injury. While most MS therapies target the immunologic response, there is a large unmet need for treatments that can promote CNS repair. LINGO-1 (leucine-rich repeat and Ig-containing Nogo receptor interacting protein-1) is a membrane protein selectively expressed in the CNS that suppresses myelination, preventing the repair of damaged axons. We are investigating LINGO-1 antagonist antibodies that lead to remyelination as a new paradigm for treatment of individuals with MS. The anti-LINGO-1 Li81 antibody,BIIB033, is currently in clinical trials and is the first MS treatment targeting CNS repair. Here, to elucidate the mechanism of action of the antibody, we solved the crystal structure of the LINGO-1-Li81 Fab complex and used biochemical and functional studies to investigate structure-function relationships. Li81 binds to the convex surface of the leucine-rich repeat domain of LINGO-1 within repeats 4-8. Fab binding blocks contact points used in the oligomerization of LINGO-1 and produces a stable complex containing two copies each of LINGO-1 and Fab that results from a rearrangement of contacts stabilizing the quaternary structure of LINGO-1. The formation of the LINGO-1-Li81 Fab complex masks functional epitopes within the Ig domain of LINGO-1 that are important for its biologic activity in oligodendrocyte differentiation. These studies provide new insights into the structure and biology of LINGO-1 and how Li81 monoclonal antibody can block its function.
Organizational Affiliation:
Departments of Drug and Molecular Discovery, Biogen Idec, Inc., Cambridge, Massachusetts blake.pepinsky@biogenidec.com.