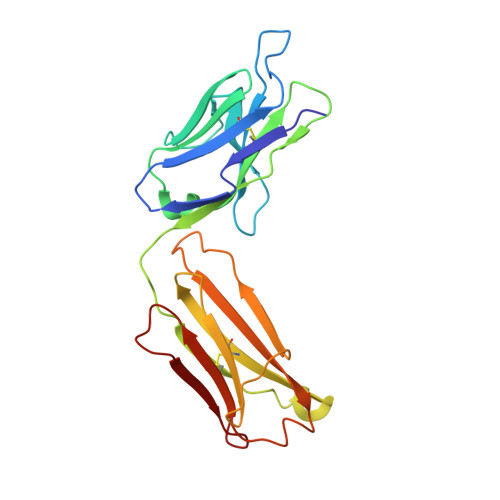
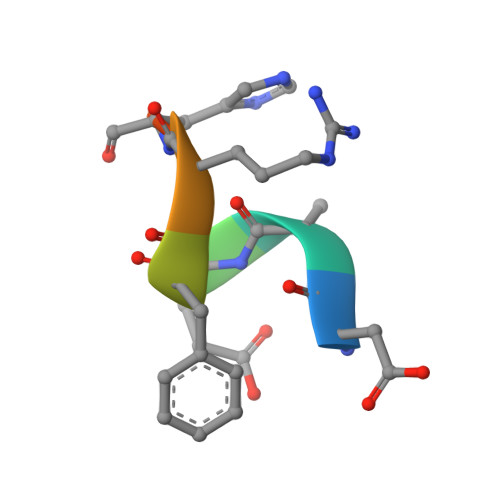
Crystallization and preliminary X-ray diffraction analysis of the Fab portion of the Alzheimer's disease immunotherapy candidate bapineuzumab complexed with amyloid-beta
Crespi, G.A.N., Ascher, D.B., Parker, M.W., Miles, L.A.(2014) Acta Crystallogr F Struct Biol Commun 70: 374-377
- PubMed: 24598931
- DOI: https://doi.org/10.1107/S2053230X14001642
- Primary Citation of Related Structures:
4OJF - PubMed Abstract:
Bapineuzumab (AAB-001) and its derivative (AAB-003) are humanized versions of the anti-Aβ murine antibody 3D6 and are immunotherapy candidates in Alzheimer's disease. The common Fab fragment of these immunotherapies has been expressed, purified and crystallized in complex with β-amyloid peptides (residues 1-8 and 1-28). Diffraction data at high resolution were acquired from crystals of Fab-Aβ8 (2.0 Å) and Fab-Aβ28 (2.2 Å) complexes at the Australian Synchrotron. Both crystal forms belonged to the primitive orthorhombic space group P21221.
Organizational Affiliation:
ACRF Rational Drug Discovery Centre and Biota Structural Biology Laboratory, St Vincent's Institute of Medical Research, 9 Princes Street, Fitzroy, Victoria 3056, Australia.