The structure of a far-red fluorescent protein, AQ143, shows evidence in support of reported red-shifting chromophore interactions.
Wannier, T.M., Mayo, S.L.(2014) Protein Sci 23: 1148-1153
- PubMed: 24888769
- DOI: https://doi.org/10.1002/pro.2498
- Primary Citation of Related Structures:
4OHS - PubMed Abstract:
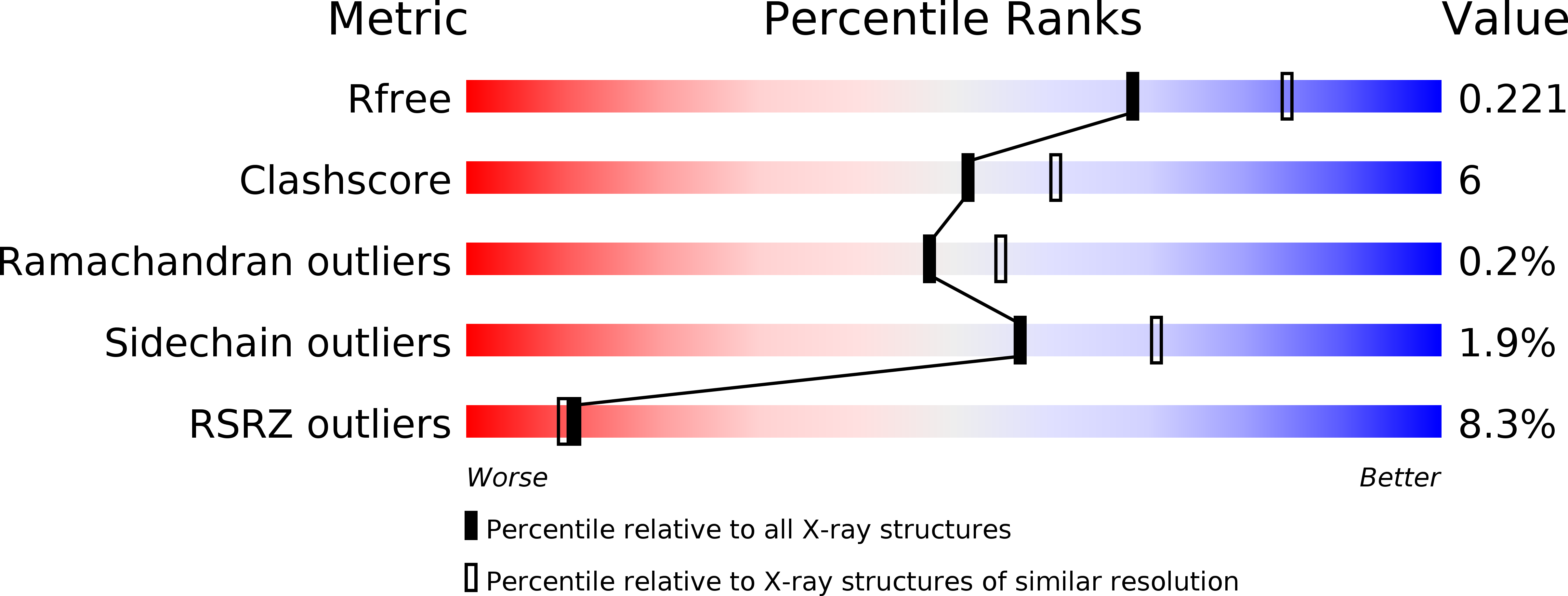
Engineering fluorescent proteins (FPs) to emit light at longer wavelengths is a significant focus in the development of the next generation of fluorescent biomarkers, as far-red light penetrates tissue with minimal absorption, allowing better imaging inside of biological hosts. Structure-guided design and directed evolution have led to the discovery of red FPs with significant bathochromic shifts to their emission. Here, we present the crystal structure of one of the most bathochromically shifted FPs reported to date, AQ143, a nine-point mutant of aeCP597, a chromoprotein from Actinia equina. The 2.19 Å resolution structure reveals several important chromophore interactions that contribute to the protein's far-red emission and shows dual occupancy of the green and red chromophores.
Organizational Affiliation:
Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, California, 91125.