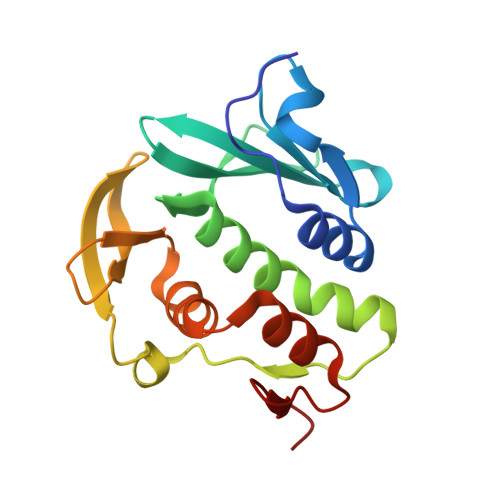
1.55 angstrom resolution X-ray crystal structure of Rv3902c from Mycobacterium tuberculosis.
Reddy, B.G., Moates, D.B., Kim, H.B., Green, T.J., Kim, C.Y., Terwilliger, T.C., Delucas, L.J.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 414-417
- PubMed: 24699730
- DOI: https://doi.org/10.1107/S2053230X14003793
- Primary Citation of Related Structures:
4O6G - PubMed Abstract:
The crystallographic structure of the Mycobacterium tuberculosis (TB) protein Rv3902c (176 residues; molecular mass of 19.8 kDa) was determined at 1.55 Å resolution. The function of Rv3902c is unknown, although several TB genes involved in bacterial pathogenesis are expressed from the operon containing the Rv3902c gene. The unique structural fold of Rv3902c contains two domains, each consisting of antiparallel β-sheets and α-helices, creating a hand-like binding motif with a small binding pocket in the palm. Structural homology searches reveal that Rv3902c has an overall structure similar to that of the Salmonella virulence-factor chaperone InvB, with an r.m.s.d. for main-chain atoms of 2.3 Å along an aligned domain.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, 1025 18th Street South, Birmingham, AL 35233, USA.