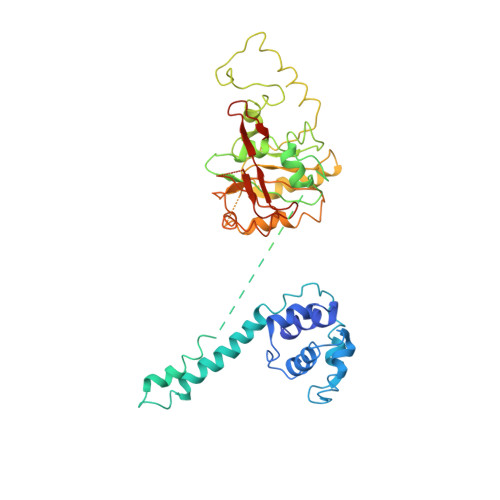
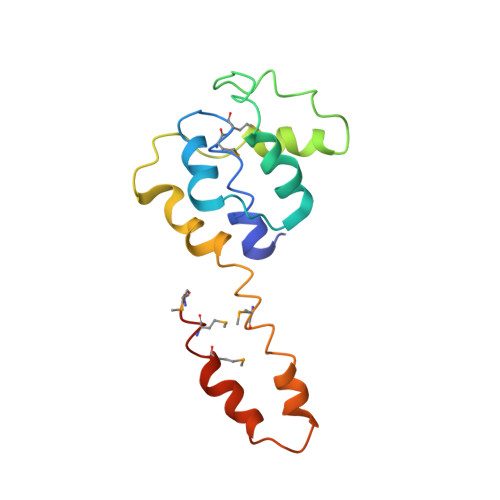
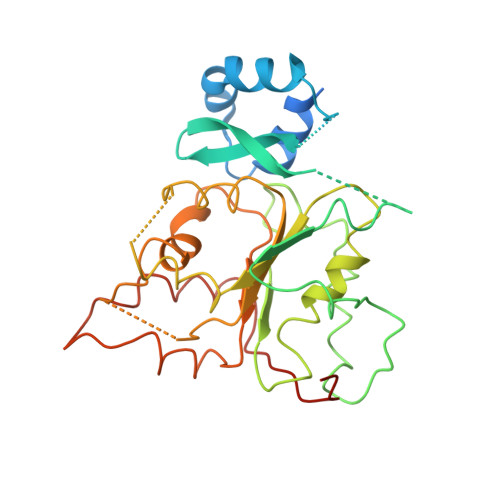
Structure of a helicase-helicase loader complex reveals insights into the mechanism of bacterial primosome assembly.
Liu, B., Eliason, W.K., Steitz, T.A.(2013) Nat Commun 4: 2495-2495
- PubMed: 24048025
- DOI: https://doi.org/10.1038/ncomms3495
- Primary Citation of Related Structures:
4M4W - PubMed Abstract:
During the assembly of the bacterial loader-dependent primosome, helicase loader proteins bind to the hexameric helicase ring, deliver it onto the oriC DNA and then dissociate from the complex. Here, to provide a better understanding of this key process, we report the crystal structure of the ~570-kDa prepriming complex between the Bacillus subtilis loader protein and the Bacillus stearothermophilus helicase, as well as the helicase-binding domain of primase with a molar ratio of 6:6:3 at 7.5 Å resolution. The overall architecture of the complex exhibits a three-layered ring conformation. Moreover, the structure combined with the proposed model suggests that the shift from the 'open-ring' to the 'open-spiral' and then the 'closed-spiral' state of the helicase ring due to the binding of single-stranded DNA may be the cause of the loader release.
Organizational Affiliation:
1] Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520, USA [2] Howard Hughes Medical Institute, New Haven, Connecticut 06510, USA.