Structural Stability of Influenza A(H1N1)pdm09 Virus Hemagglutinins.
Yang, H., Chang, J.C., Guo, Z., Carney, P.J., Shore, D.A., Donis, R.O., Cox, N.J., Villanueva, J.M., Klimov, A.I., Stevens, J.(2014) J Virol 88: 4828-4838
- PubMed: 24522930
- DOI: https://doi.org/10.1128/JVI.02278-13
- Primary Citation of Related Structures:
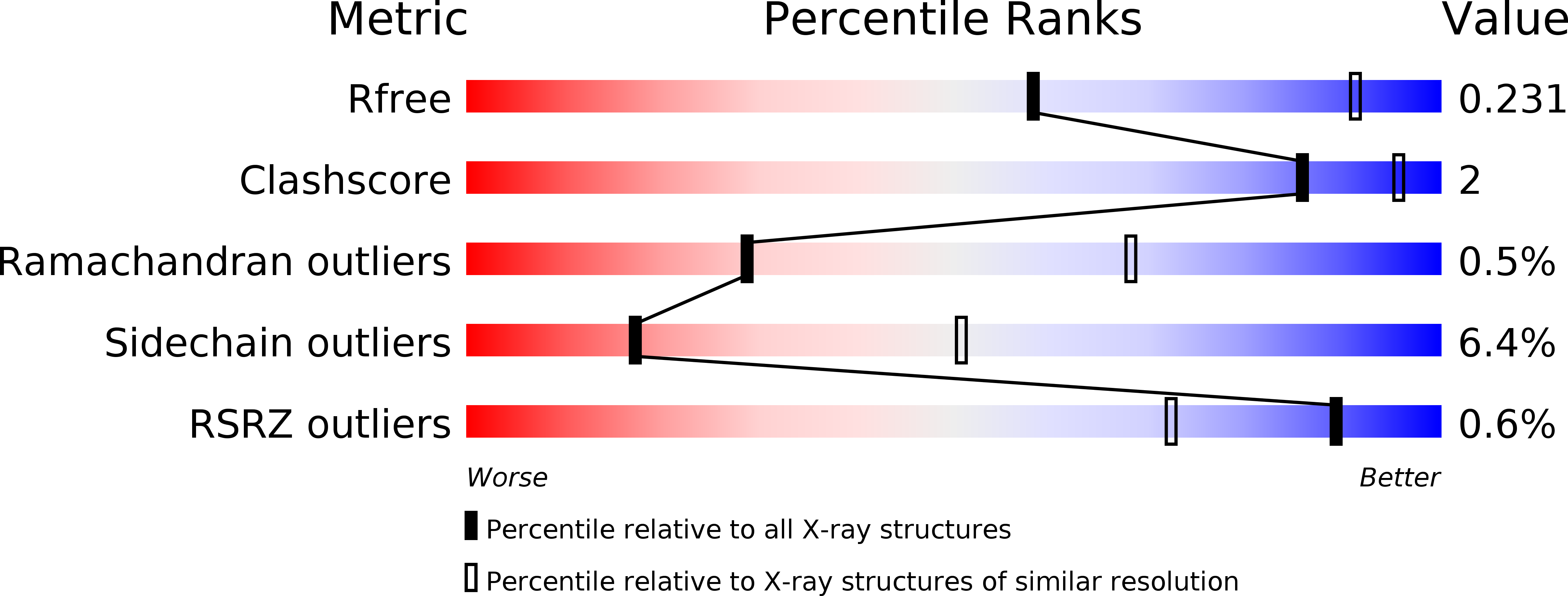
4LXV - PubMed Abstract:
The noncovalent interactions that mediate trimerization of the influenza hemagglutinin (HA) are important determinants of its biological activities. Recent studies have demonstrated that mutations in the HA trimer interface affect the thermal and pH sensitivities of HA, suggesting a possible impact on vaccine stability (). We used size exclusion chromatography analysis of recombinant HA ectodomain to compare the differences among recombinant trimeric HA proteins from early 2009 pandemic H1N1 viruses, which dissociate to monomers, with those of more recent virus HAs that can be expressed as trimers. We analyzed differences among the HA sequences and identified intermolecular interactions mediated by the residue at position 374 (HA0 numbering) of the HA2 subdomain as critical for HA trimer stability. Crystallographic analyses of HA from the recent H1N1 virus A/Washington/5/2011 highlight the structural basis for this observed phenotype. It remains to be seen whether more recent viruses with this mutation will yield more stable vaccines in the future. Hemagglutinins from the early 2009 H1N1 pandemic viruses are unable to maintain a trimeric complex when expressed in a recombinant system. However, HAs from 2010 and 2011 strains are more stable, and our work highlights that the improvement in stability can be attributed to an E374K substitution in the HA2 subunit of the stalk that emerged naturally in the circulating viruses.
Organizational Affiliation:
Influenza Division, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.