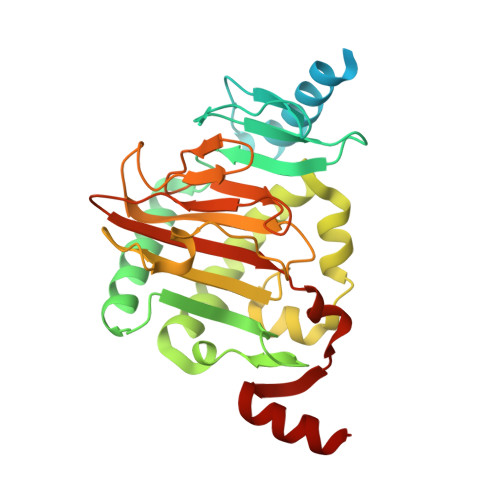
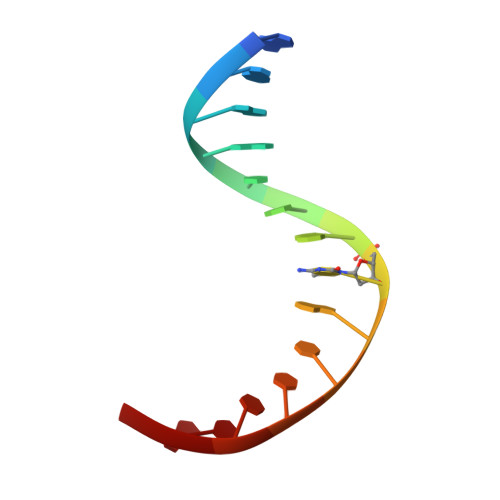
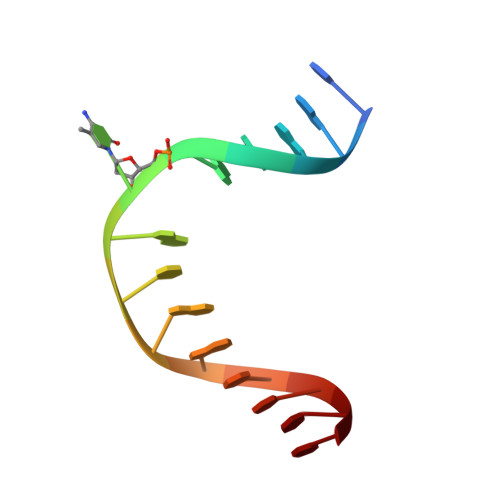
Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Hashimoto, H., Pais, J.E., Zhang, X., Saleh, L., Fu, Z.Q., Dai, N., Correa, I.R., Zheng, Y., Cheng, X.(2014) Nature 506: 391-395
- PubMed: 24390346
- DOI: https://doi.org/10.1038/nature12905
- Primary Citation of Related Structures:
4LT5 - PubMed Abstract:
Cytosine residues in mammalian DNA occur in five forms: cytosine (C), 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). The ten-eleven translocation (Tet) dioxygenases convert 5mC to 5hmC, 5fC and 5caC in three consecutive, Fe(II)- and α-ketoglutarate-dependent oxidation reactions. The Tet family of dioxygenases is widely distributed across the tree of life, including in the heterolobosean amoeboflagellate Naegleria gruberi. The genome of Naegleria encodes homologues of mammalian DNA methyltransferase and Tet proteins. Here we study biochemically and structurally one of the Naegleria Tet-like proteins (NgTet1), which shares significant sequence conservation (approximately 14% identity or 39% similarity) with mammalian Tet1. Like mammalian Tet proteins, NgTet1 acts on 5mC and generates 5hmC, 5fC and 5caC. The crystal structure of NgTet1 in complex with DNA containing a 5mCpG site revealed that NgTet1 uses a base-flipping mechanism to access 5mC. The DNA is contacted from the minor groove and bent towards the major groove. The flipped 5mC is positioned in the active-site pocket with planar stacking contacts, Watson-Crick polar hydrogen bonds and van der Waals interactions specific for 5mC. The sequence conservation between NgTet1 and mammalian Tet1, including residues involved in structural integrity and functional significance, suggests structural conservation across phyla.
Organizational Affiliation:
Departments of Biochemistry, Emory University School of Medicine, 1510 Clifton Road, Atlanta, Georgia 30322, USA.