Structural evidence for a two-regime photobleaching mechanism in a reversibly switchable fluorescent protein.
Duan, C., Adam, V., Byrdin, M., Ridard, J., Kieffer-Jaquinod, S., Morlot, C., Arcizet, D., Demachy, I., Bourgeois, D.(2013) J Am Chem Soc 135: 15841-15850
- PubMed: 24059326
- DOI: https://doi.org/10.1021/ja406860e
- Primary Citation of Related Structures:
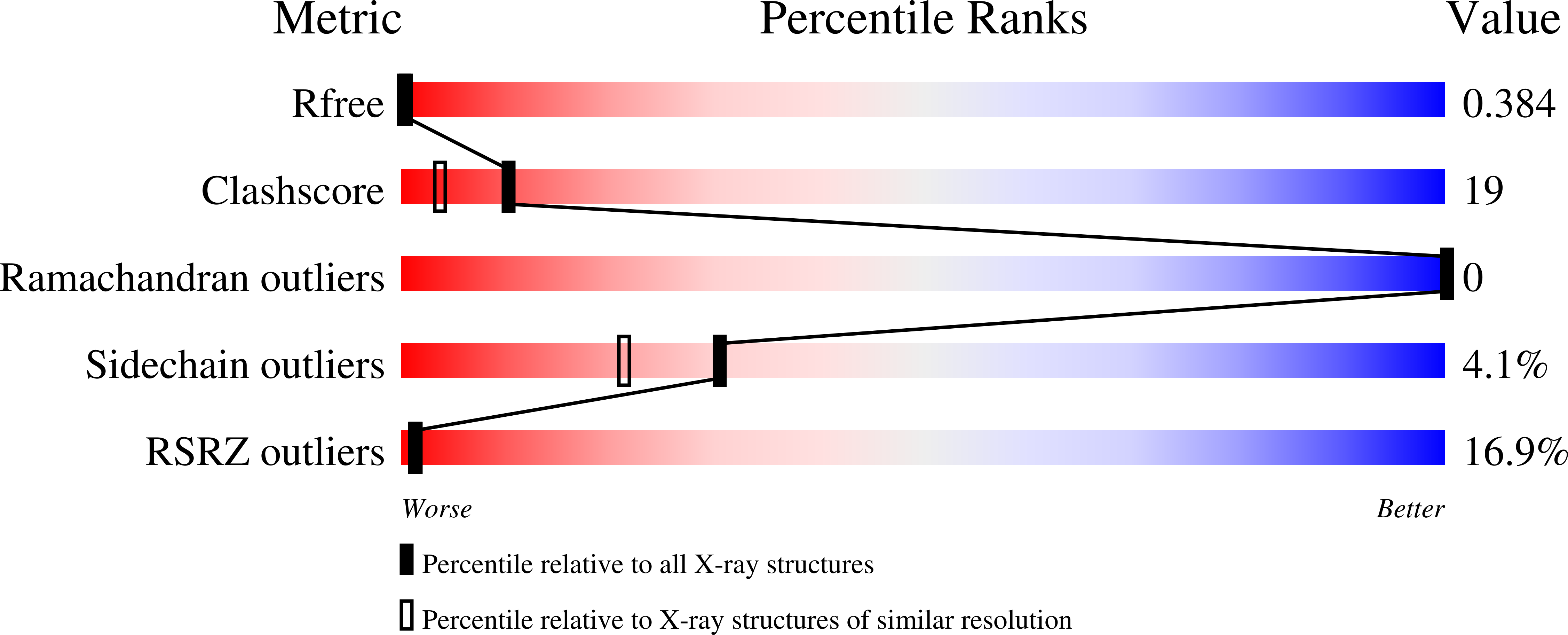
4LJB, 4LJC, 4LJD - PubMed Abstract:
Photobleaching, the irreversible photodestruction of a chromophore, severely limits the use of fluorescent proteins (FPs) in optical microscopy. Yet, the mechanisms that govern photobleaching remain poorly understood. In Reversibly Switchable Fluorescent Proteins (RSFPs), a class of FPs that can be repeatedly photoswitched between nonfluorescent and fluorescent states, photobleaching limits the achievable number of switching cycles, a process known as photofatigue. We investigated the photofatigue mechanisms in the protein IrisFP using combined X-ray crystallography, optical in crystallo spectroscopy, mass spectrometry and modeling approaches. At laser-light intensities typical of conventional wide-field fluorescence microscopy, an oxygen-dependent photobleaching pathway was evidenced. Structural modifications induced by singlet-oxygen production within the chromophore pocket revealed the oxidation of two sulfur-containing residues, Met159 and Cys171, locking the chromophore in a nonfluorescent protonated state. At laser-light intensities typical of localization-based nanoscopy (>0.1 kW/cm(2)), a completely different, oxygen-independent photobleaching pathway was found to take place. The conserved Glu212 underwent decarboxylation concomitantly with an extensive rearrangement of the H-bond network around the chromophore, and an sp(2)-to-sp(3) hybridization change of the carbon atom bridging the chromophore cyclic moieties was observed. This two-regime photobleaching mechanism is likely to be a common feature in RSFPs from Anthozoan species, which typically share high structural and sequence identity with IrisFP. In addition, our results suggest that, when such FPs are used, the illumination conditions employed in localization-based super-resolution microscopy might generate less cytotoxicity than those of standard wide-field microscopy at constant absorbed light-dose. Finally, our data will facilitate the rational design of FPs displaying enhanced photoresistance.
Organizational Affiliation:
Université Grenoble Alpes , Institut de Biologie Structurale (IBS), F-38027 Grenoble, France.