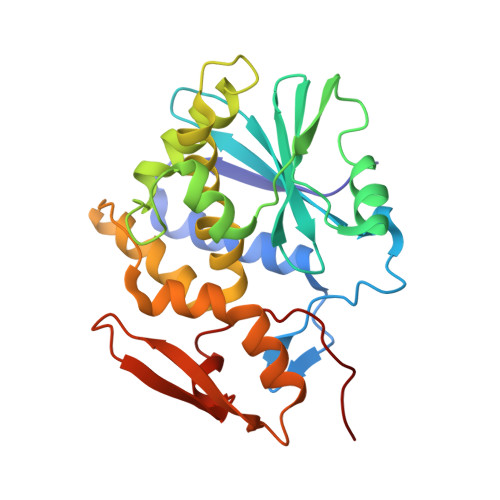
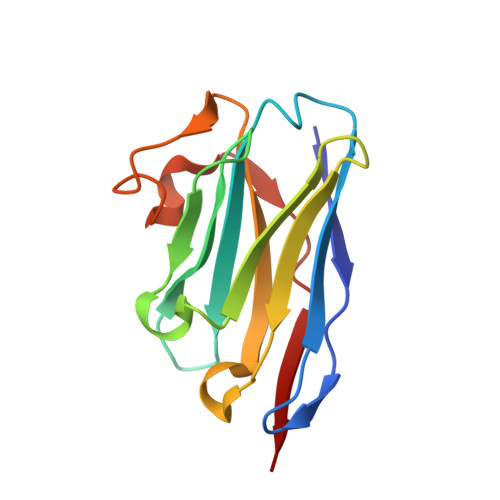
Crystal Structures of Ricin Toxin's Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies.
Rudolph, M.J., Vance, D.J., Cheung, J., Franklin, M.C., Burshteyn, F., Cassidy, M.S., Gary, E.N., Herrera, C., Shoemaker, C.B., Mantis, N.J.(2014) J Mol Biol 426: 3057-3068
- PubMed: 24907552
- DOI: https://doi.org/10.1016/j.jmb.2014.05.026
- Primary Citation of Related Structures:
4LGP, 4LGR, 4LGS, 4LHJ, 4LHQ - PubMed Abstract:
Ricin is a select agent toxin and a member of the RNA N-glycosidase family of medically important plant and bacterial ribosome-inactivating proteins. In this study, we determined X-ray crystal structures of the enzymatic subunit of ricin (RTA) in complex with the antigen binding domains (VHH) of five unique single-chain monoclonal antibodies that differ in their respective toxin-neutralizing activities. None of the VHHs made direct contact with residues involved in RTA's RNA N-glycosidase activity or induced notable allosteric changes in the toxin's subunit. Rather, the five VHHs had overlapping structural epitopes on the surface of the toxin and differed in the degree to which they made contact with prominent structural elements in two folding domains of the RTA. In general, RTA interactions were influenced most by the VHH CDR3 (CDR, complementarity-determining region) elements, with the most potent neutralizing antibody having the shortest and most conformationally constrained CDR3. These structures provide unique insights into the mechanisms underlying toxin neutralization and provide critically important information required for the rational design of ricin toxin subunit vaccines.
Organizational Affiliation:
New York Structural Biology Center, New York, NY 10027, USA. Electronic address: mrudolph@nysbc.org.