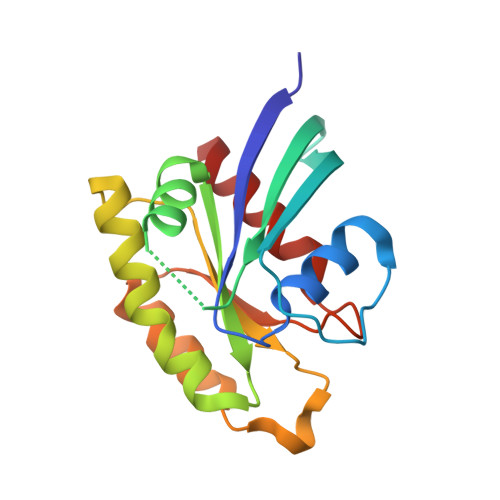
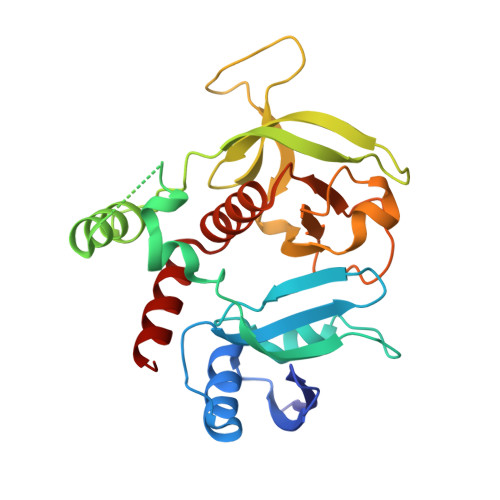
The structure of Rap1 in complex with RIAM reveals specificity determinants and recruitment mechanism.
Zhang, H., Chang, Y.C., Brennan, M.L., Wu, J.(2014) J Mol Cell Biol 6: 128-139
- PubMed: 24287201
- DOI: https://doi.org/10.1093/jmcb/mjt044
- Primary Citation of Related Structures:
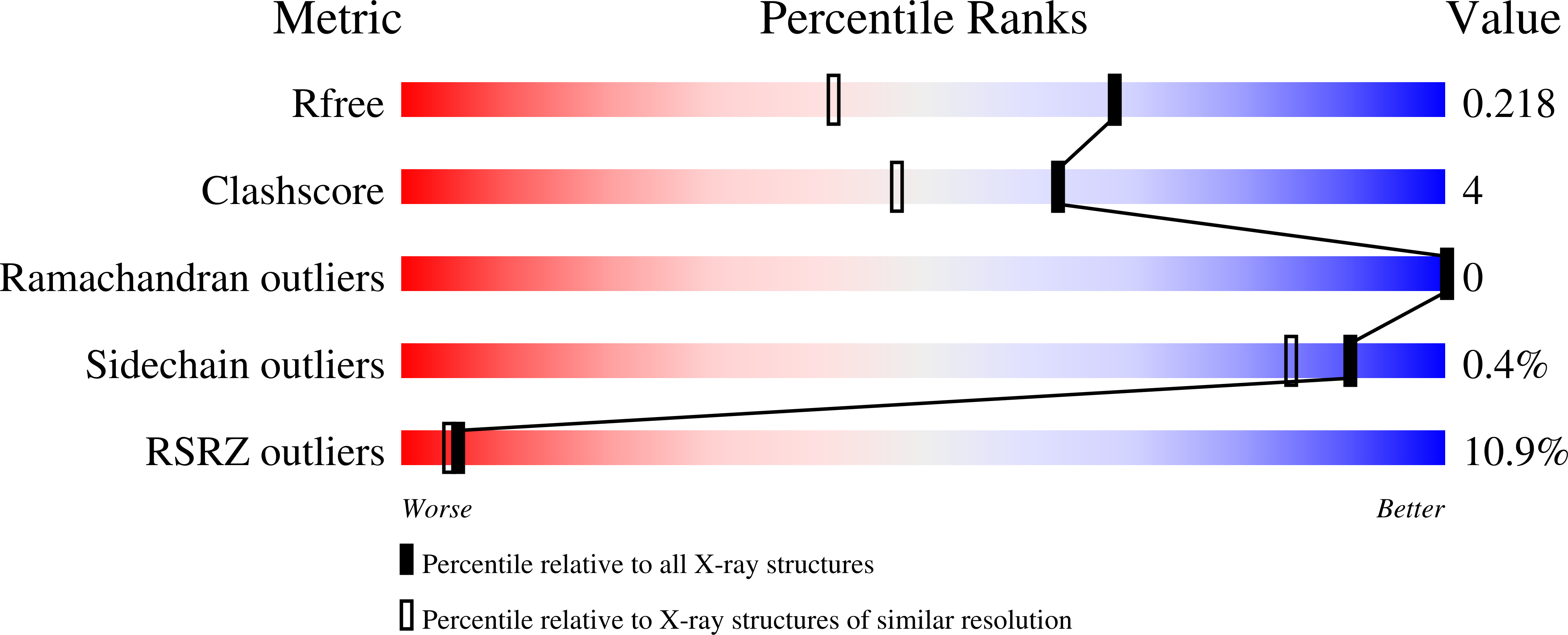
4KVG - PubMed Abstract:
The small GTPase Rap1 induces integrin activation via an inside-out signaling pathway mediated by the Rap1-interacting adaptor molecule (RIAM). Blocking this pathway may suppress tumor metastasis and other diseases that are related to hyperactive integrins. However, the molecular basis for the specific recognition of RIAM by Rap1 remains largely unknown. Herein we present the crystal structure of an active, GTP-bound GTPase domain of Rap1 in complex with the Ras association (RA)-pleckstrin homology (PH) structural module of RIAM at 1.65 Å. The structure reveals that the recognition of RIAM by Rap1 is governed by side-chain interactions. Several side chains are critical in determining specificity of this recognition, particularly the Lys31 residue in Rap1 that is oppositely charged compared with the Glu31/Asp31 residue in other Ras GTPases. Lys31 forms a salt bridge with RIAM residue Glu212, making it the key specificity determinant of the interaction. We also show that disruption of these interactions results in reduction of Rap1:RIAM association, leading to a loss of co-clustering and cell adhesion. Our findings elucidate the molecular mechanism by which RIAM mediates Rap1-induced integrin activation. The crystal structure also offers new insight into the structural basis for the specific recruitment of RA-PH module-containing effector proteins by their small GTPase partners.
Organizational Affiliation:
Developmental Therapeutics Program, Fox Chase Cancer Center, 333 Cottman Avenue, Philadelphia, PA 19111, USA.