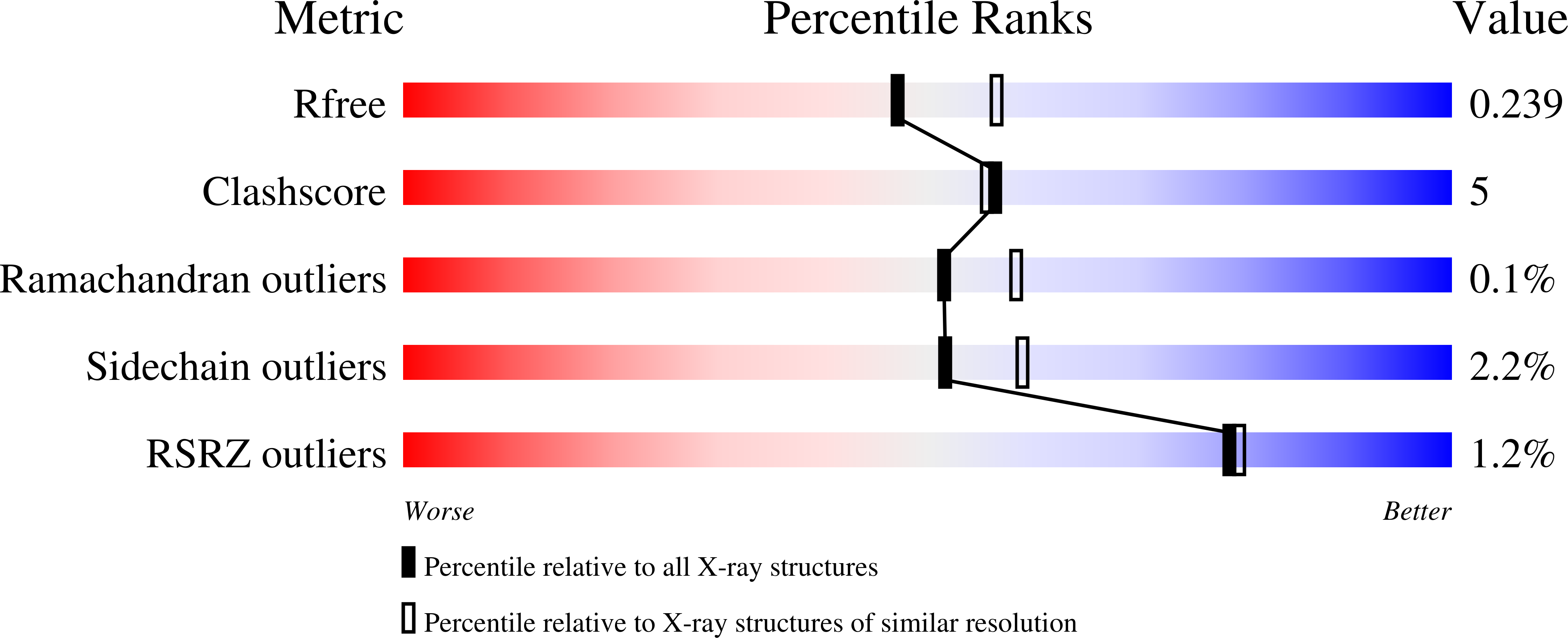
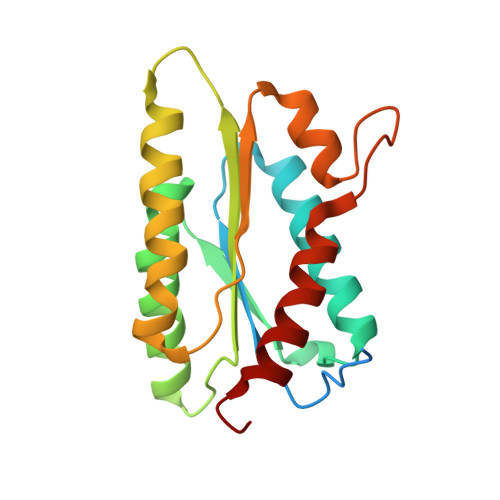
Catalysis product captured in lumazine synthase from the fungal pathogen Candida glabrata.
Shankar, M., Wilbanks, S.M., Nakatani, Y., Monk, B.C., Tyndall, J.D.(2013) Acta Crystallogr D Biol Crystallogr 69: 1580-1586
- PubMed: 23897480
- DOI: https://doi.org/10.1107/S0907444913010949
- Primary Citation of Related Structures:
4KQ6 - PubMed Abstract:
Candida glabrata has emerged as an important fungal pathogen with intrinsic resistance to azole drugs. The limited efficacy of and resistance to existing antifungals is driving the need to identify new drug targets. The enzyme 6,7-dimethyl-8-(D-ribityl)lumazine synthase is part of the riboflavin-biosynthesis pathway essential to fungi and bacteria and is a potential drug target for the development of broad-spectrum antifungal drugs. The X-ray crystal structure of recombinant lumazine synthase from C. glabrata was obtained at 2.24 Å resolution and revealed a dimer of homopentamers, with one in five subunits containing a product molecule from the catalytic reaction.
Organizational Affiliation:
School of Pharmacy, University of Otago, PO Box 56, Dunedin 9054, New Zealand.