The rotational order-disorder structure of the reversibly photoswitchable red fluorescent protein rsTagRFP.
Pletnev, S., Subach, F.V., Verkhusha, V.V., Dauter, Z.(2014) Acta Crystallogr D Biol Crystallogr 70: 31-39
- PubMed: 24419376
- DOI: https://doi.org/10.1107/S1399004713024644
- Primary Citation of Related Structures:
4KPI - PubMed Abstract:
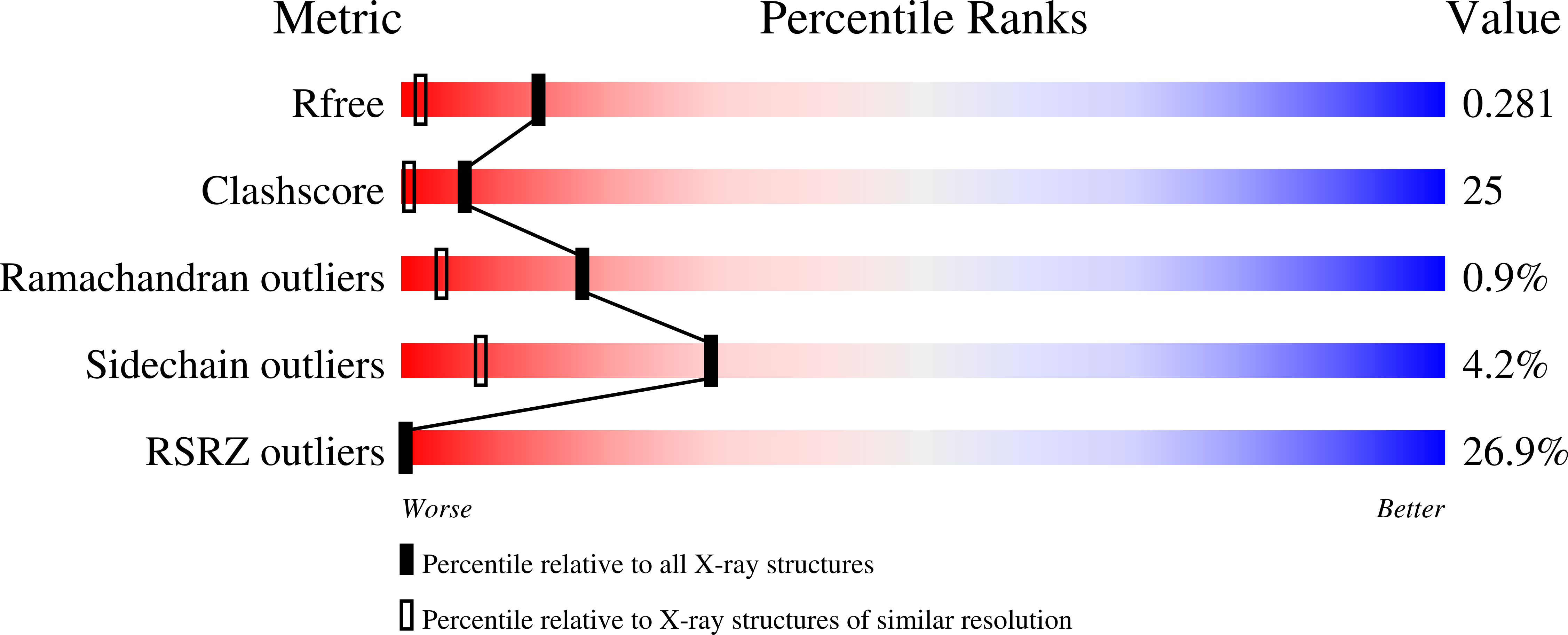
The rotational order-disorder (OD) structure of the reversibly photoswitchable fluorescent protein rsTagRFP is discussed in detail. The structure is composed of tetramers of 222 symmetry incorporated into the lattice in two different orientations rotated 90° with respect to each other around the crystal c axis and with tetramer axes coinciding with the crystallographic twofold axes. The random distribution of alternatively oriented tetramers in the crystal creates the rotational OD structure with statistically averaged I422 symmetry. Despite order-disorder pathology, the structure of rsTagRFP has electron-density maps of good quality for both non-overlapping and overlapping parts of the model. The crystal contacts, crystal internal architecture and a possible mechanism of rotational OD crystal formation are discussed.
Organizational Affiliation:
Leidos Biomedical Research Inc., Basic Research Program, Argonne National Laboratory, Argonne, IL 60439, USA.