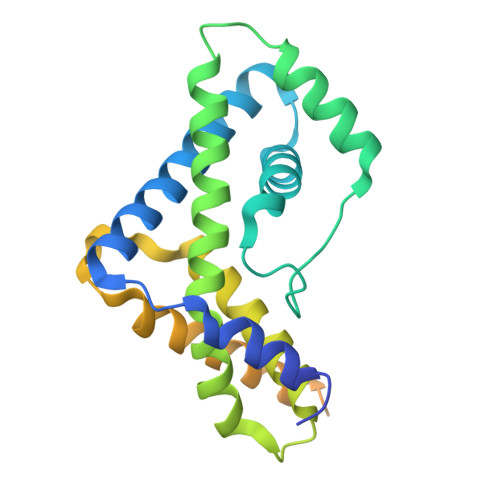
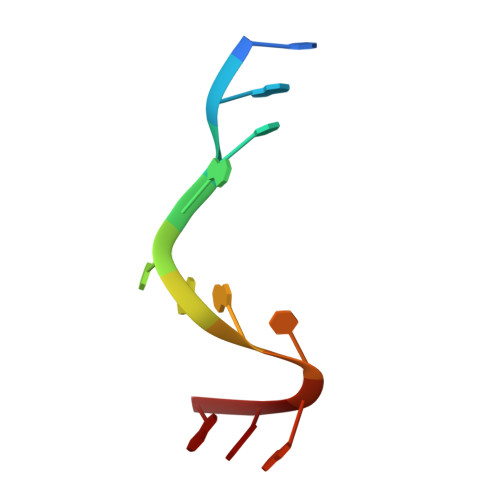Crystallographic analysis of an RNA polymerase sigma-subunit fragment complexed with -10 promoter element ssDNA: quadruplex formation as a possible tool for engineering crystal contacts in protein-ssDNA complexes.
Feklistov, A., Darst, S.A.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 950-955
- PubMed: 23989139
- DOI: https://doi.org/10.1107/S1744309113020368
- Primary Citation of Related Structures:
4KI2 - PubMed Abstract:
Structural studies of -10 promoter element recognition by domain 2 of the RNA polymerase σ subunit [Feklistov & Darst (2011), Cell, 147, 1257-1269] reveal an unusual crystal-packing arrangement dominated by G-quartets. The 3'-terminal GGG motif of the oligonucleotide used in crystallization participates in G-quadruplex formation with GGG motifs from symmetry-related complexes. Stacking between neighboring G-quadruplexes results in the formation of pseudo-continuous four-stranded columns running throughout the length of the crystal (G-columns). Here, a new crystal form is presented with a different arrangement of G-columns and it is proposed that the fortuitous finding of G-quartet packing could be useful in engineering crystal contacts in protein-ssDNA complexes.
Organizational Affiliation:
Laboratory of Molecular Biophysics, The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA. afeklistov@rockefeller.edu