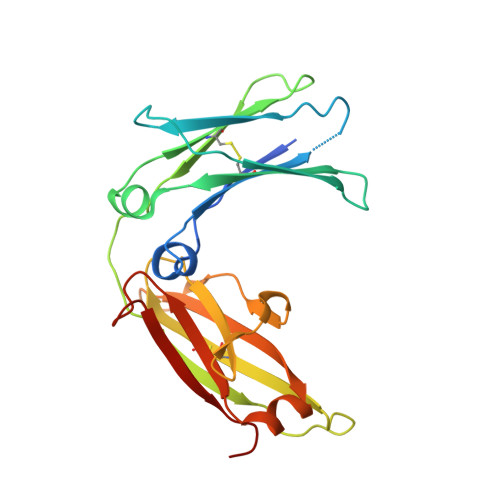
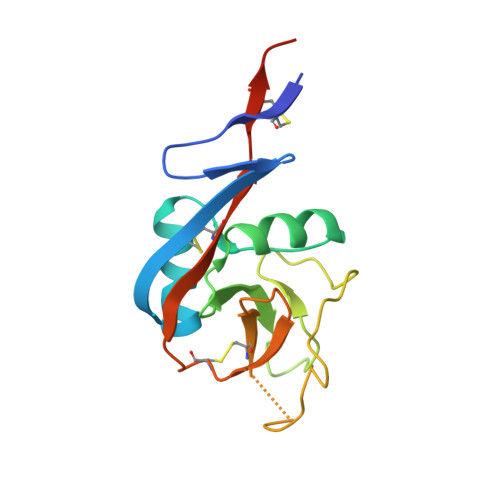
A range of C3-C4 interdomain angles in IgE Fc accommodate binding to its receptor CD23.
Dhaliwal, B., Pang, M.O., Yuan, D., Beavil, A.J., Sutton, B.J.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 305-309
- PubMed: 24598915
- DOI: https://doi.org/10.1107/S2053230X14003355
- Primary Citation of Related Structures:
4KI1 - PubMed Abstract:
The antibody IgE plays a central role in allergic disease, functioning principally through two cell-surface receptors: FcℇRI and CD23. FcℇRI on mast cells and basophils mediates the immediate hypersensitivity response, whilst the interaction of IgE with CD23 on B cells regulates IgE production. Crystal structures of the lectin-like `head' domain of CD23 alone and bound to a subfragment of IgE consisting of the dimer of Cℇ3 and Cℇ4 domains (Fcℇ3-4) have recently been determined, revealing flexibility in the IgE-binding site of CD23. Here, a new crystal form of the CD23-Fcℇ3-4 complex with different molecular-packing constraints is reported, which together with the earlier results demonstrates that conformational variability at the interface extends additionally to the IgE Fc and the quaternary structure of its domains.
Organizational Affiliation:
Randall Division of Cell and Molecular Biophysics, King's College London, New Hunt's House, Guy's Campus, London SE1 1UL, England.