Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
Bianchetti, C.M., Harmann, C.H., Takasuka, T.E., Hura, G.L., Dyer, K., Fox, B.G.(2013) J Biol Chem 288: 18574-18587
- PubMed: 23653358
- DOI: https://doi.org/10.1074/jbc.M113.475848
- Primary Citation of Related Structures:
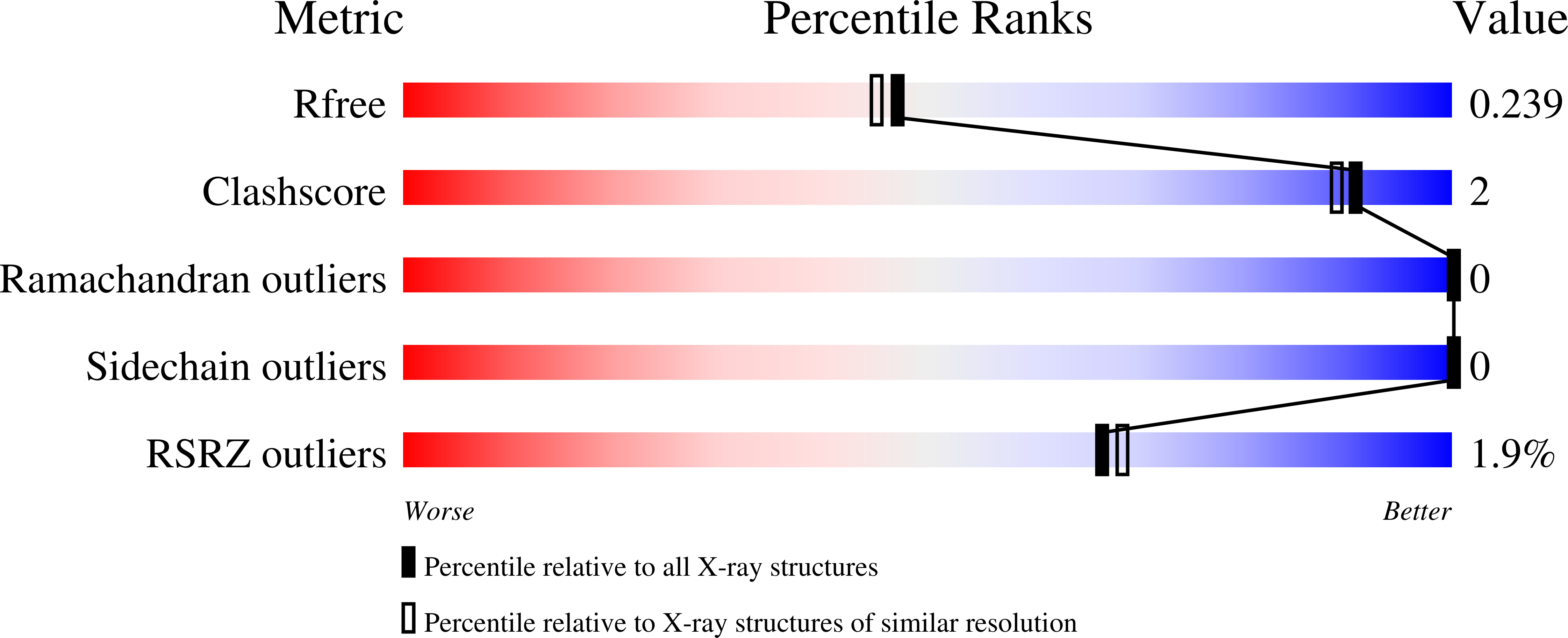
4ILT, 4ILV - PubMed Abstract:
Streptomyces sp. SirexAA-E is a highly cellulolytic bacterium isolated from an insect/microbe symbiotic community. When grown on lignin-containing biomass, it secretes SACTE_2871, an aromatic ring dioxygenase domain fused to a family 5/12 carbohydrate-binding module (CBM 5/12). Here we present structural and catalytic studies of this novel fusion enzyme, thus providing insight into its function. The dioxygenase domain has the core β-sandwich fold typical of this enzyme family but lacks a dimerization domain observed in other intradiol dioxygenases. Consequently, the x-ray structure shows that the enzyme is monomeric and the Fe(III)-containing active site is exposed to solvent in a shallow depression on a planar surface. Purified SACTE_2871 catalyzes the O2-dependent intradiol cleavage of catechyl compounds from lignin biosynthetic pathways, but not their methylated derivatives. Binding studies show that SACTE_2871 binds synthetic lignin polymers and chitin through the interactions of the CBM 5/12 domain, representing a new binding specificity for this fold-family. Based on its unique structural features and functional properties, we propose that SACTE_2871 contributes to the invasive nature of the insect/microbial community by destroying precursors needed by the plant for de novo lignin biosynthesis as part of its natural wounding response.
Organizational Affiliation:
Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, Madison, Wisconsin, 53706, USA.