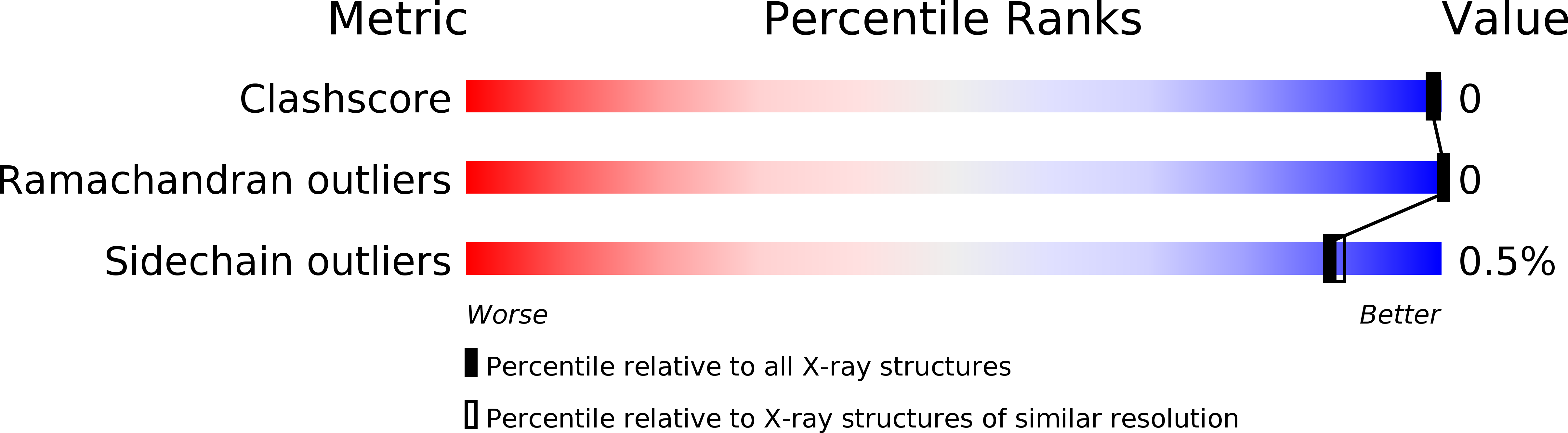Neutron and X-ray crystallographic analysis of Achromobacter protease I at pD 8.0: protonation states and hydration structure in the free-form.
Ohnishi, Y., Yamada, T., Kurihara, K., Tanaka, I., Sakiyama, F., Masaki, T., Niimura, N.(2013) Biochim Biophys Acta 1834: 1642-1647
- PubMed: 23714114
- DOI: https://doi.org/10.1016/j.bbapap.2013.05.012
- Primary Citation of Related Structures:
4GPG - PubMed Abstract:
The structure of the free-form of Achromobacter protease I (API) at pD 8.0 was refined by simultaneous use of single crystal X-ray and neutron diffraction data sets to investigate the protonation states of key catalytic residues of the serine protease. Occupancy refinement of the catalytic triad in the active site of API free-form showed that ca. 30% of the imidazole ring of H57 and ca. 70% of the hydroxyl group of S194 were deuterated. This observation indicates that a major fraction of S194 is protonated in the absence of a substrate. The protonation state of the catalytic triad in API was compared with the bovine β-trypsin-BPTI complex. The comparison led to the hypothesis that close contact of a substrate with S194 could lower the acidity of its hydroxyl group, thereby allowing H57 to extract the hydrogen from the hydroxyl group of S194. H210, which is a residue specific to API, does not form a hydrogen bond with the catalytic triad residue D113. Instead, H210 forms a hydrogen bond network with S176, H177 and a water molecule. The close proximity of the bulky, hydrophobic residue W169 may protect this hydrogen bond network, and this protection may stabilize the function of API over a wide pH range.
Organizational Affiliation:
Faculty of Engineering, Ibaraki University, Hitachi, Ibaraki, Japan.