An Ultra-specific Avian Antibody to Phosphorylated Tau Protein Reveals a Unique Mechanism for Phosphoepitope Recognition.
Shih, H.H., Tu, C., Cao, W., Klein, A., Ramsey, R., Fennell, B.J., Lambert, M., Ni Shuilleabhain, D., Autin, B., Kouranova, E., Laxmanan, S., Braithwaite, S., Wu, L., Ait-Zahra, M., Milici, A.J., Dumin, J.A., Lavallie, E.R., Arai, M., Corcoran, C., Paulsen, J.E., Gill, D., Cunningham, O., Bard, J., Mosyak, L., Finlay, W.J.(2012) J Biol Chem 287: 44425-44434
- PubMed: 23148212
- DOI: https://doi.org/10.1074/jbc.M112.415935
- Primary Citation of Related Structures:
4GLR - PubMed Abstract:
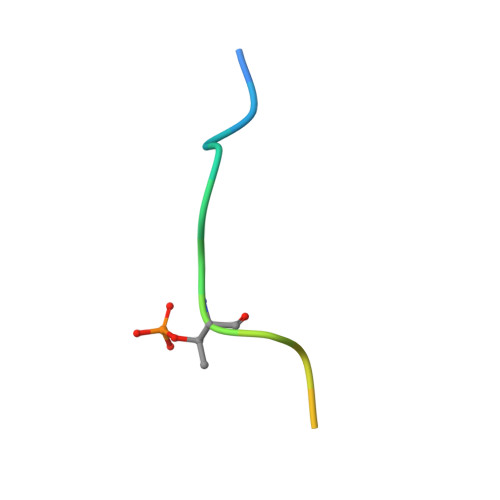
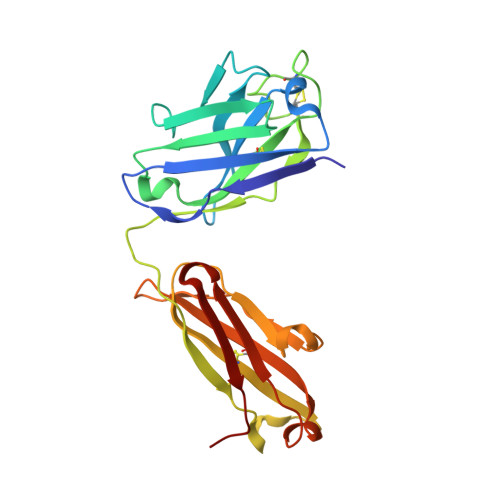
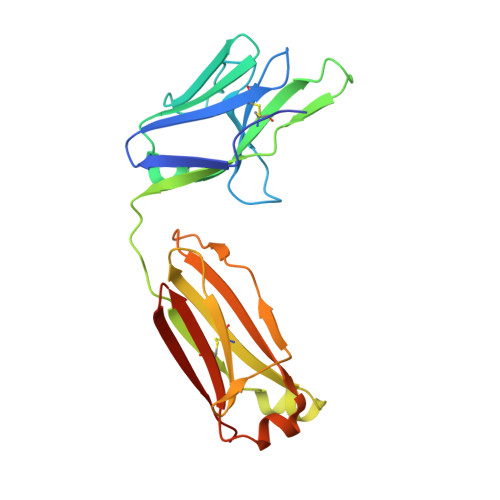
Highly specific antibodies to phosphoepitopes are valuable tools to study phosphorylation in disease states, but their discovery is largely empirical, and the molecular mechanisms mediating phosphospecific binding are poorly understood. Here, we report the generation and characterization of extremely specific recombinant chicken antibodies to three phosphoepitopes on the Alzheimer disease-associated protein tau. Each antibody shows full specificity for a single phosphopeptide. The chimeric IgG pT231/pS235_1 exhibits a K(D) of 0.35 nm in 1:1 binding to its cognate phosphopeptide. This IgG is murine ortholog-cross-reactive, specifically recognizing the pathological form of tau in brain samples from Alzheimer patients and a mouse model of tauopathy. To better understand the underlying binding mechanisms allowing such remarkable specificity, we determined the structure of pT231/pS235_1 Fab in complex with its cognate phosphopeptide at 1.9 Å resolution. The Fab fragment exhibits novel complementarity determining region (CDR) structures with a "bowl-like" conformation in CDR-H2 that tightly and specifically interacts with the phospho-Thr-231 phosphate group, as well as a long, disulfide-constrained CDR-H3 that mediates peptide recognition. This binding mechanism differs distinctly from either peptide- or hapten-specific antibodies described to date. Surface plasmon resonance analyses showed that pT231/pS235_1 binds a truly compound epitope, as neither phosphorylated Ser-235 nor free peptide shows any measurable binding affinity.
Organizational Affiliation:
Global Biotherapeutics Technologies, Pfizer Global Research & Development, Cambridge, Massachusetts 02140, USA.