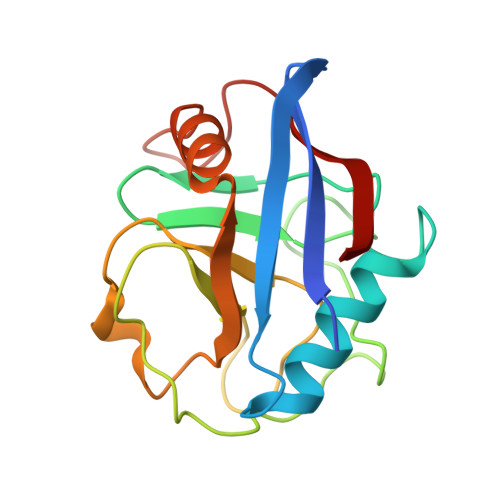
Structure of RNA-interacting Cyclophilin A-like protein from Piriformospora indica that provides salinity-stress tolerance in plants
Trivedi, D.K., Bhatt, H., Pal, R.K., Tuteja, R., Garg, B., Johri, A.K., Bhavesh, N.S., Tuteja, N.(2013) Sci Rep 3: 3001-3001
- PubMed: 24141523
- DOI: https://doi.org/10.1038/srep03001
- Primary Citation of Related Structures:
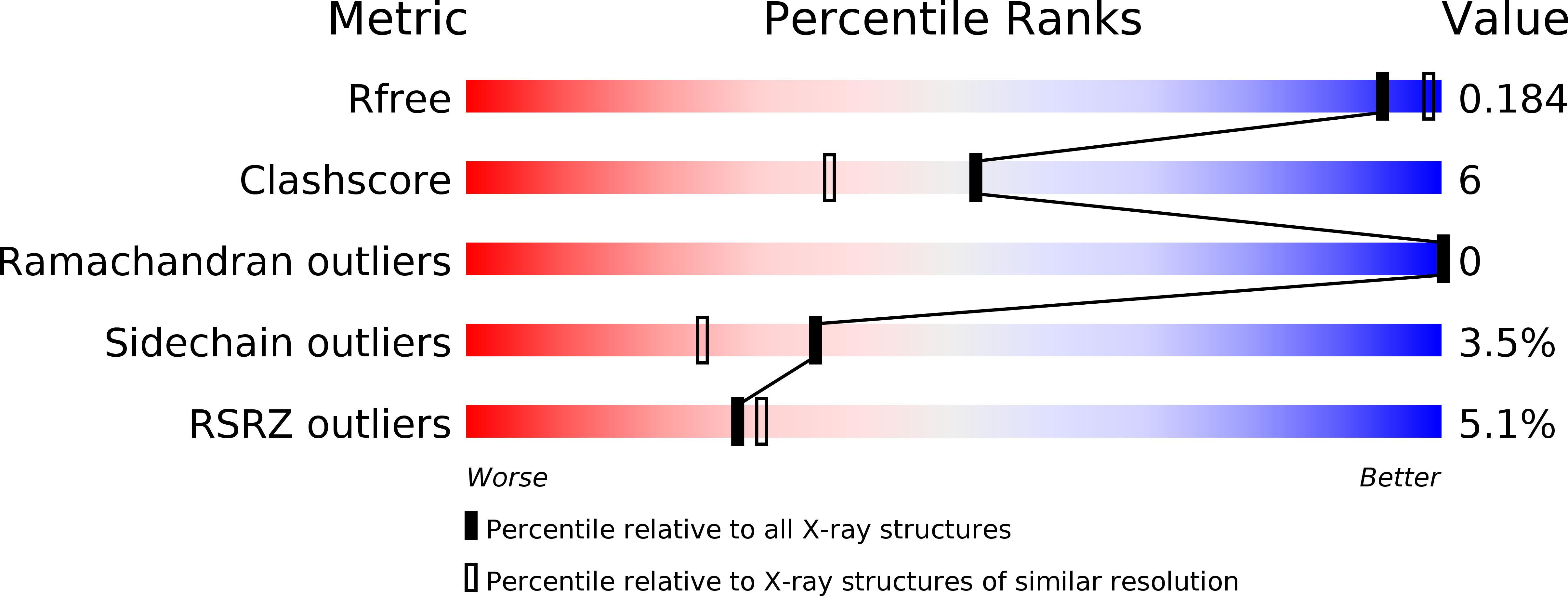
4EYV - PubMed Abstract:
Soil salinity problems are widespread around the globe with increased risk of spreading over the years. The fungus Piriformospora indica, identified in Indian Thar desert, colonizes the roots of monocotyledon plants and provides resistance towards biotic as well as abiotic stress conditions. We have identified a cyclophilin A-like protein from P. indica (PiCypA), which shows higher expression levels during salinity stress. The transgenic tobacco plants overexpressing PiCypA develop osmotic tolerance and exhibit normal growth under osmotic stress conditions. The crystal structure and NMR spectroscopy of PiCypA show a canonical cyclophilin like fold exhibiting a novel RNA binding activity. The RNA binding activity of the protein and identification of the key residues involved in the RNA recognition is unique for this class of protein. Here, we demonstrate for the first time a direct evidence of countering osmotic stress tolerance in plant by genetic modification using a P. indica gene.
Organizational Affiliation:
1] Plant Molecular Biology group [2].