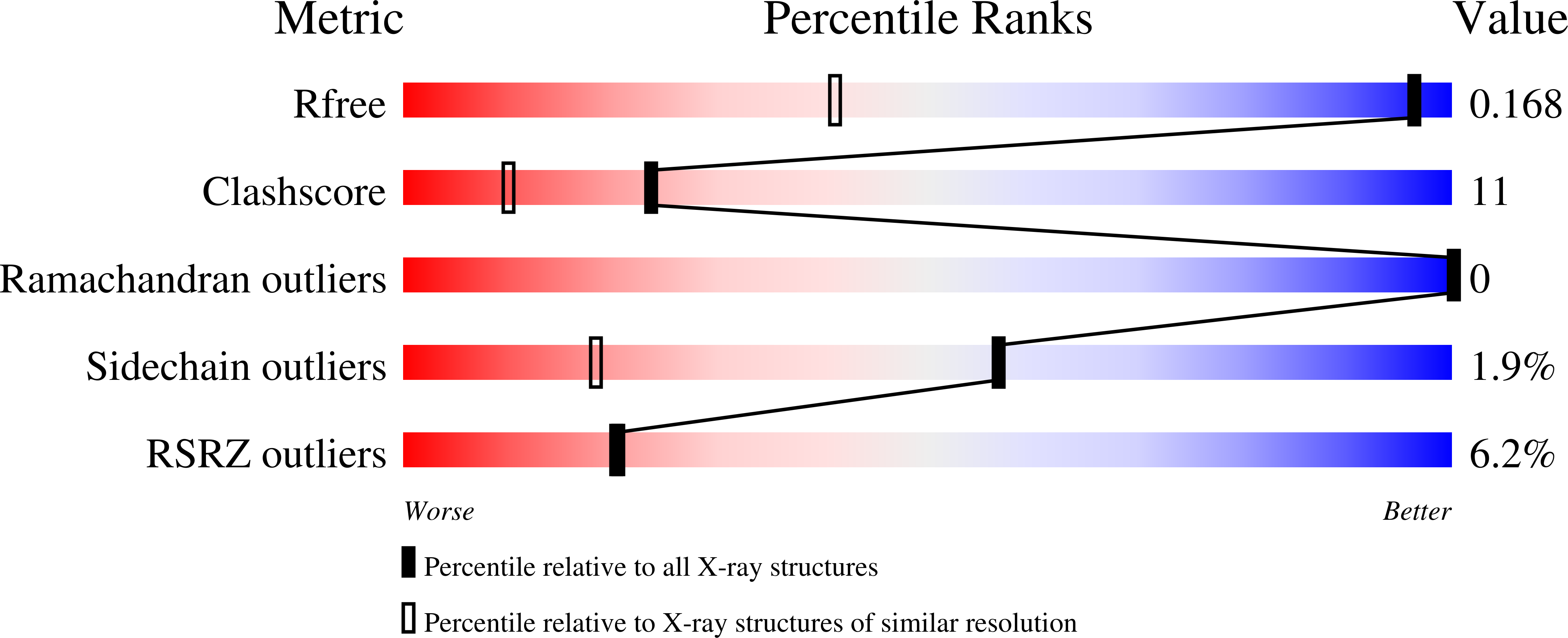
Crystal Structure of Peanut ( Arachis hypogaea ) Allergen Ara h 5.
Wang, Y., Fu, T.J., Howard, A., Kothary, M.H., McHugh, T.H., Zhang, Y.(2013) J Agric Food Chem 61: 1573-1578
- PubMed: 23350842
- DOI: https://doi.org/10.1021/jf303861p
- Primary Citation of Related Structures:
4ESP - PubMed Abstract:
Profilins from numerous species are known to be allergens, including food allergens, such as peanut ( Arachis hypogaea ) allergen Ara h 5, and pollen allergens, such as birch allergen Bet v 2. Patients with pollen allergy can also cross-react to peanut. Structural characterization of allergens will allow a better understanding of the allergenicity of food allergens and their cross-reactivities. The three-dimensional structures of most known food allergens remain to be elucidated. Here, we report the first crystallographic study of a food allergen in the profilin family. The structure of peanut allergen Ara h 5 was determined, and the resolution of the final refined structure was 1.1 Å. Structure alignment revealed that Ara h 5 is more similar to Bet v 2 than to Hev b 8, although sequence alignment suggested that Ara h 5 is more closely related to Hev b 8 than to Bet v 2, indicating that homology-model-based prediction of immunoglobulin E epitopes needs to be interpreted with caution.
Organizational Affiliation:
Department of Biological and Chemical Sciences, Illinois Institute of Technology, Chicago, Illinois 60616, United States.