Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing modified pyrimidines
Obeid, S., Bu kamp, H., Welte, W., Diederichs, K., Marx, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA polymerase I, thermostable | 540 | Thermus aquaticus | Mutation(s): 0 Gene Names: polA, pol1 EC: 2.7.7.7 | 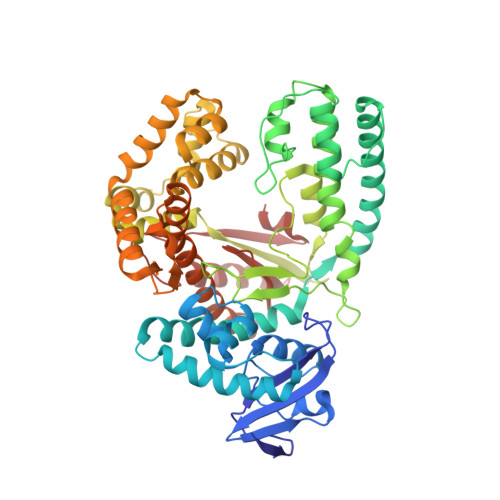 | |
UniProt | |||||
Find proteins for P19821 (Thermus aquaticus) Explore P19821 Go to UniProtKB: P19821 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P19821 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(0R8))-3') | 12 | synthetic construct | 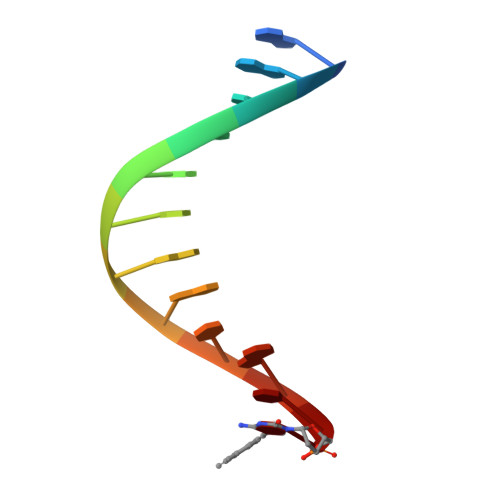 | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*AP*AP*AP*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3') | 16 | synthetic construct | 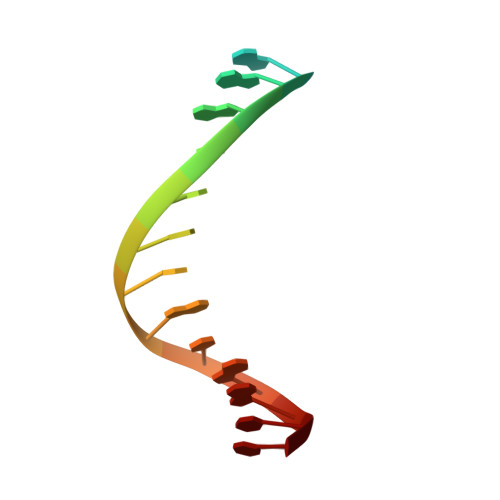 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 0R7 Query on 0R7 | E [auth A] | [[(2S,5R)-5-[4-azanyl-5-[2-(4-ethynylphenyl)ethynyl]-2-oxidanylidene-pyrimidin-1-yl]oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate C19 H20 N3 O12 P3 PROFTMDIPXHDEG-DLBZAZTESA-N |  | ||
| PGE Query on PGE | G [auth A] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | F [auth A], H [auth A], I [auth A], M [auth C] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | J [auth A], O [auth C] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CA Query on CA | K [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| MG Query on MG | D [auth A], L [auth B], N [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 109.213 | α = 90 |
| b = 109.213 | β = 90 |
| c = 91.047 | γ = 120 |
| Software Name | Purpose |
|---|---|
| XDS | data scaling |
| PHENIX | model building |
| PHENIX | refinement |
| XDS | data reduction |
| PHENIX | phasing |